Clustering and viewing GSEA results
c_gsea.Rmd
library(EnrichGT)
#> Loading required package: dplyr
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
#> Loading required package: tibble
#> Loading required package: gt
#> Loading required package: cli
#>
#> ── EnrichGT ────────────────────────────────────────────────────────────────────
#> ℹ View your enrichment result by entring `EnrichGT(result)`
#> → by Zhiming Ye, https://github.com/ZhimingYe/EnrichGTSummary
Similar to ORA, for GSEA enrichment analysis (What is GSEA enrichment
analysis? See webpage: https://yulab-smu.top/biomedical-knowledge-mining-book/enrichment-overview.html),
EnrichGT can read results from gseaResult generated by
clusterProfiler or an exported table from clusterProfiler
enrichment (including at least the columns
“ID”,“Description”,“NES”,“pvalue”,“p.adjust” and “core_enrichment”.
EnrichGT performs term frequency statistics and automatic
clustering based on genes enriched in hits, returns an S4 object, and
outputs a beautifully formatted result wrapped by gt.
Parsing GSEA results
This only shows results. For detailing usage, please see the Article
of ORA (the before article).
We encourage run clusterProfiler::setReadable() first
before passing to EnrichGT(), to get gene symbols correctly
(this is a bad example without setReadable()).
suppressMessages({
library(tidyverse)
library(gt)
library(clusterProfiler)
library(org.Hs.eg.db)
})
data(geneList, package="DOSE")
ego3 <- gseGO(geneList = geneList,
OrgDb = org.Hs.eg.db,
ont = "CC",
minGSSize = 100,
maxGSSize = 500,
pvalueCutoff = 0.05,
verbose = FALSE)
#> Warning in fgseaMultilevel(pathways = pathways, stats = stats, minSize =
#> minSize, : For some pathways, in reality P-values are less than 1e-10. You can
#> set the `eps` argument to zero for better estimation.
(EnrichGT(ego3, ClusterNum = 100, P.adj = 1)@gt_object)
#> Loading required package: text2vec
#>
#> Attaching package: 'text2vec'
#> The following object is masked from 'package:BiocGenerics':
#>
#> normalize
#> ℹ Please run setReadable first!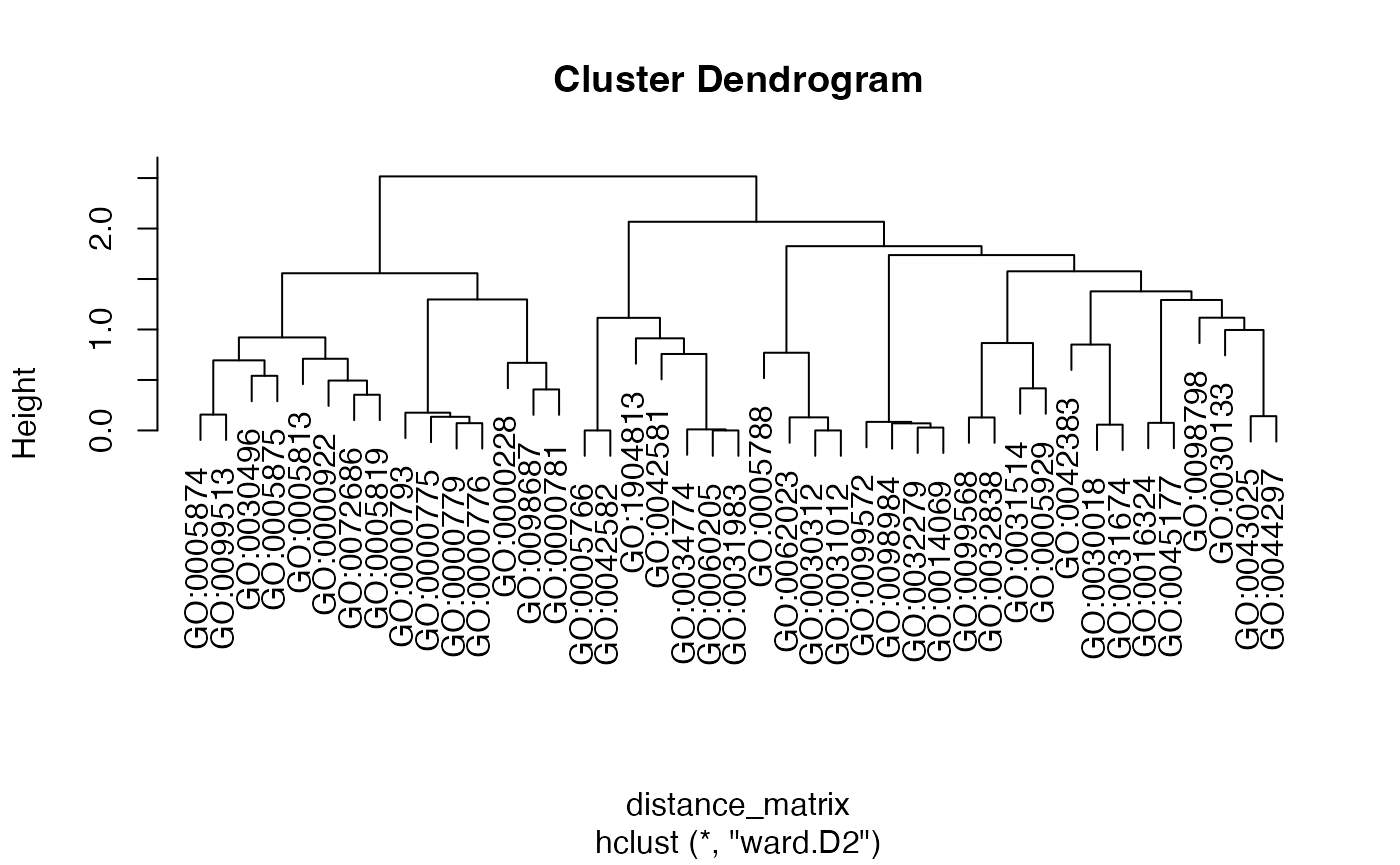
| Parse form: ego3 | ||||
| Split into 3 Clusters. Generated by github@zhimingye/EnrichGT | ||||
| Description | absNES | Padj | core_enrichment | |
|---|---|---|---|---|
| Cluster_1 | ||||
|
chromosome, centromeric region
GO:0000775
|
2.718 | 1.4e-09 | 55143, 991, 1062, 10403, 7153, 55355, 220134, 4751, 79019, 55839, 54821, 4085, 81930, 81620, 332, 2146, 7272, 64151, 9212, 6790, 891, 11004, 5347, 701, 11130, 79682, 57405, 10615, 79075, 2491, 11339, 3070, 9918, 1058, 699, 1063, 55055, 1051, 8970, 79980, 9735, 55732, 23310 | |
|
condensed chromosome, centromeric region
GO:0000779
|
2.713 | 1.4e-09 | 991, 1062, 10403, 55355, 220134, 4751, 79019, 55839, 54821, 4085, 81930, 81620, 332, 7272, 64151, 9212, 6790, 891, 11004, 5347, 701, 11130, 79682, 57405, 10615, 2491, 9918, 1058, 699, 1063, 55055, 1051, 79980, 9735, 55732, 23310 | |
|
kinetochore
GO:0000776
|
2.670 | 1.4e-09 | 991, 1062, 10403, 55355, 220134, 4751, 79019, 55839, 54821, 4085, 81930, 81620, 332, 7272, 9212, 6790, 891, 11004, 5347, 701, 11130, 79682, 57405, 10615, 2491, 699, 1063, 55055, 79980, 9735, 55732 | |
|
chromosomal region
GO:0098687
|
2.584 | 1.4e-09 | 55143, 991, 1062, 10403, 7153, 55355, 220134, 4751, 79019, 10635, 55839, 983, 54821, 4085, 81930, 81620, 332, 2146, 7272, 64151, 9212, 1111, 6790, 891, 4174, 4171, 11004, 5347, 701, 11130, 79682, 57405, 10615, 79075, 2491, 5888, 4998, 11339, 3070, 4175, 4173, 2237, 9918, 1058, 699, 1063, 5111, 9401, 55055, 55506, 641, 1763, 1051, 8970, 4176, 79980, 4436, 9735, 55732, 23310, 3066, 5499, 10051, 4172, 5424, 5885, 11200, 2072, 92822, 54908, 6396, 4683, 54069, 86, 3018, 1788, 142, 6839, 23028, 1786, 3014, 5905, 3619, 11335, 55166, 54107, 54433, 7517 | |
|
nuclear chromosome
GO:0000228
|
2.584 | 1.4e-09 | 8318, 55388, 7153, 23397, 4751, 9837, 332, 64151, 51659, 1111, 4174, 4171, 5347, 5888, 23594, 4998, 4175, 4173, 54962, 9918, 84296, 81611, 5111, 64785, 55506, 641, 54892, 5427, 23649, 4176, 58516, 5557, 23310, 3066, 11169, 8607, 10051, 1104, 5558, 4172, 5424, 5885, 7283, 10592, 8914, 51377, 86, 5393, 142, 6839, 23212, 3014, 3619, 5425, 54107, 11177, 7273, 6119 | |
|
condensed chromosome
GO:0000793
|
2.538 | 1.4e-09 | 991, 1062, 10403, 7153, 23397, 55355, 220134, 4751, 79019, 55839, 54821, 4085, 81930, 81620, 332, 7272, 64151, 9212, 1111, 6790, 891, 11004, 5347, 701, 11130, 79682, 57405, 10615, 2491, 5888, 4288, 9918, 1058, 699, 1063, 55055, 641, 1051, 54892, 3148, 79980, 9735, 55732, 23310, 10051, 1104 | |
|
mitotic spindle
GO:0072686
|
2.348 | 1.4e-09 | 9493, 1062, 259266, 9787, 220134, 51203, 22974, 10460, 983, 4085, 81930, 3832, 50852, 9212, 9055, 3833, 146909, 6790, 990, 5347, 29127, 10615, 1894, 9700, 54801, 54959, 29899, 65263 | |
|
spindle
GO:0005819
|
2.224 | 1.4e-09 | 55143, 991, 9493, 1062, 259266, 9787, 220134, 51203, 22974, 10460, 4751, 983, 4085, 81930, 332, 3832, 50852, 7272, 9212, 9055, 3833, 146909, 10112, 6790, 891, 24137, 9928, 3161, 11004, 79801, 990, 5347, 29127, 701, 10615, 1894, 9700, 56992, 54801, 54959, 29899, 994, 65263, 1063, 26271, 55055, 29901, 8317, 79980, 9585, 9735, 55732, 292, 5778, 4064, 284403, 79866, 5885, 203068, 55856, 57553, 79000, 84722, 51115, 7283, 10381, 51647 | |
|
midbody
GO:0030496
|
2.105 | 6.1e-08 | 55143, 9493, 1062, 259266, 55165, 4751, 983, 332, 9212, 9055, 10112, 6790, 24137, 9928, 79801, 5347, 29127, 10615, 1894 | |
|
microtubule
GO:0005874
|
1.708 | 8.3e-06 | 9493, 1062, 259266, 220134, 51203, 22974, 4751, 983, 81930, 332, 3832, 9212, 9055, 3833, 146909, 10112, 6790, 24137, 9928, 11004, 5347, 29127, 51512, 10615, 56992, 54801, 4599, 26150, 22948, 9654, 10397, 55055, 79739, 79745, 3925, 7277, 27285, 9585, 9735, 908, 29105, 80086, 10576, 4600, 84790, 8514, 10263, 203068, 10059, 84722, 55346, 51115, 7283, 10381 | |
| Cluster_2 | ||||
|
collagen-containing extracellular matrix
GO:0062023
|
2.164 | 1.4e-09 | 4017, 6649, 1288, 4811, 3910, 51162, 3371, 2662, 12, 1291, 1301, 7474, 3490, 79875, 7450, 80781, 78987, 1490, 1280, 1306, 80070, 8425, 977, 10085, 4054, 4256, 7482, 7837, 7042, 3912, 5919, 2817, 8722, 6694, 1278, 10216, 1511, 5176, 4060, 283, 30008, 7869, 1277, 7078, 5549, 22795, 10516, 81578, 23452, 1293, 1295, 22915, 8406, 80760, 1012, 6469, 8076, 5118, 2192, 1281, 10218, 50509, 1290, 81029, 79812, 7058, 11096, 2202, 4313, 2199, 1294, 4856, 4653, 6387, 11117, 3263, 3339, 1462, 1289, 1292, 3908, 4016, 25878, 3909, 4053, 6678, 1955, 1296, 7041, 8292, 633, 1191, 2162, 10418, 2239, 1842, 165, 5654, 10631, 1805, 81035, 2331, 4485, 2152, 63923, 7043, 5104, 1359, 3913, 8404, 1300, 2200, 1634, 131578, 4059, 7177, 1287, 563, 7060, 2006, 7373, 1307, 9370, 8416, 79148, 1311, 1308, 8483, 4693, 4148, 64499, 54829, 4239, 4969 | |
|
external encapsulating structure
GO:0030312
|
2.105 | 1.4e-09 | 57124, 3910, 51162, 3371, 2662, 12, 1291, 1301, 56547, 7474, 5016, 3490, 79875, 7450, 80781, 78987, 79625, 8840, 1490, 1280, 1306, 4314, 80070, 8425, 977, 10085, 4054, 4256, 3483, 7482, 2246, 7837, 169611, 80059, 7042, 3912, 4322, 5919, 2817, 8722, 6694, 1278, 10216, 1511, 5176, 4060, 283, 30008, 7869, 1277, 7078, 5549, 22795, 10516, 81578, 23452, 1293, 1295, 4588, 22915, 8406, 80760, 1012, 6469, 8076, 5118, 2192, 1281, 10218, 50509, 4319, 1290, 81029, 25903, 79812, 7058, 11096, 2202, 4313, 2199, 1294, 4856, 26018, 7481, 4653, 6387, 11117, 3263, 3339, 7079, 1462, 1289, 1292, 3908, 4016, 25878, 3909, 4053, 83716, 6678, 1955, 1296, 7041, 8292, 633, 1191, 54361, 2162, 10418, 2239, 1842, 2615, 165, 5654, 10631, 1805, 81035, 2331, 4485, 3730, 54674, 2152, 63923, 7043, 25992, 5104, 1359, 3913, 4982, 8404, 1300, 2200, 1634, 131578, 4059, 7177, 1287, 563, 7060, 11081, 2006, 7373, 1307, 9370, 8857, 8416, 79148, 4958, 1311, 1308, 8483, 8839, 4693, 4148, 1101, 64499, 54829, 10234, 4239, 4969 | |
|
extracellular matrix
GO:0031012
|
2.105 | 1.4e-09 | 57124, 3910, 51162, 3371, 2662, 12, 1291, 1301, 56547, 7474, 5016, 3490, 79875, 7450, 80781, 78987, 79625, 8840, 1490, 1280, 1306, 4314, 80070, 8425, 977, 10085, 4054, 4256, 3483, 7482, 2246, 7837, 169611, 80059, 7042, 3912, 4322, 5919, 2817, 8722, 6694, 1278, 10216, 1511, 5176, 4060, 283, 30008, 7869, 1277, 7078, 5549, 22795, 10516, 81578, 23452, 1293, 1295, 4588, 22915, 8406, 80760, 1012, 6469, 8076, 5118, 2192, 1281, 10218, 50509, 4319, 1290, 81029, 25903, 79812, 7058, 11096, 2202, 4313, 2199, 1294, 4856, 26018, 7481, 4653, 6387, 11117, 3263, 3339, 7079, 1462, 1289, 1292, 3908, 4016, 25878, 3909, 4053, 83716, 6678, 1955, 1296, 7041, 8292, 633, 1191, 54361, 2162, 10418, 2239, 1842, 2615, 165, 5654, 10631, 1805, 81035, 2331, 4485, 3730, 54674, 2152, 63923, 7043, 25992, 5104, 1359, 3913, 4982, 8404, 1300, 2200, 1634, 131578, 4059, 7177, 1287, 563, 7060, 11081, 2006, 7373, 1307, 9370, 8857, 8416, 79148, 4958, 1311, 1308, 8483, 8839, 4693, 4148, 1101, 64499, 54829, 10234, 4239, 4969 | |
|
endoplasmic reticulum lumen
GO:0005788
|
1.684 | 1.1e-04 | 64374, 845, 3371, 23621, 1291, 1301, 375056, 2098, 7474, 2322, 7466, 5624, 3490, 80781, 79642, 1280, 1306, 10272, 844, 11001, 590, 3912, 3270, 6694, 1278, 56605, 80267, 51280, 414, 1277, 81578, 1293, 1295, 6469, 1281, 50509, 11167, 1290, 81029, 11096, 56034, 91851, 1294, 2621, 1471, 1462, 1289, 1292, 1296, 174, 1191, 54361, 213, 10418, 11098, 54587, 3913, 3487, 8404, 1300, 2200, 1287, 7373, 80310, 1307, 1308, 652, 4148, 8614 | |
|
mitochondrial protein-containing complex
GO:0098798
|
1.574 | 1.6e-03 | 5163, 51373, 29088, 28998, 1537, 9377, 6182, 51650, 292, 51642, 6183, 56993, 3028, 4726, 4704, 10105, 1737, 51081, 28977, 29796, 10440, 7386, 3419, 10469, 64960, 5160, 2395, 54927, 11222, 10452, 79135, 56922, 4725, 518, 4723, 9551, 506, 1349, 4708, 90480, 100287932, 4700, 509, 3421, 55168, 65003, 54539, 51069, 25874, 1351, 5161, 55052, 1329, 1678, 8050, 65005, 55750, 9868, 10245, 4719, 25813, 55669, 63875, 4697, 4702, 9801, 7818, 26521, 516, 4694, 1340, 10845, 9553, 26517, 29074, 51073, 5428, 6389, 51318, 4712, 6832, 54148, 4722, 55013, 64963, 51116, 4711, 11331, 522, 1337, 7381, 4710, 65993, 7384, 65080, 10476, 514, 581, 7416, 6150, 51025, 1327, 3396, 51021, 9167 | |
|
cytoplasmic region
GO:0099568
|
1.638 | 2.0e-03 | 1639, 51332, 79882, 23025, 55717, 4130, 50632, 146754, 3315, 10947, 29922, 6416, 1499, 56171, 9371, 25790, 10497, 92104, 27445, 2735, 25876, 5813, 6787, 5590, 2169, 66008, 5311, 22863, 23122, 51626, 4747, 9742, 6812, 5874, 51314, 4926, 22832, 582, 79819, 22906, 23116, 79659, 138162, 55812, 2737, 3800, 55638, 83450, 9576, 114327, 4117, 55259, 4137, 7802 | |
|
sarcolemma
GO:0042383
|
1.691 | 2.6e-03 | 783, 5532, 3880, 6443, 5530, 6326, 10211, 825, 6548, 1291, 7402, 7779, 5592, 8082, 2318, 80024, 844, 8910, 79026, 6717, 3752, 481, 1293, 358, 477, 27092, 113146, 6444, 6253, 9499, 3480, 6833, 857, 1292, 775, 3908, 81493, 633, 7043, 5348, 776 | |
|
plasma membrane bounded cell projection cytoplasm
GO:0032838
|
1.607 | 3.4e-03 | 55717, 4130, 50632, 146754, 3315, 10947, 29922, 6416, 56171, 9371, 25790, 92104, 2735, 25876, 5813, 6787, 66008, 22863, 51626, 4747, 9742, 5874, 51314, 22832, 582, 79819, 22906, 23116, 79659, 138162, 55812, 2737, 3800, 55638, 83450, 9576, 114327, 4117, 55259, 4137, 7802 | |
|
motile cilium
GO:0031514
|
1.566 | 7.1e-03 | 56171, 56912, 64064, 25790, 80705, 2712, 25876, 6337, 8452, 11057, 54558, 55779, 23639, 79781, 1235, 5311, 151011, 8100, 481, 51626, 51314, 582, 57728, 55064, 79819, 585, 79659, 83450, 9576, 1811, 8382, 2947, 4117, 79846, 7802 | |
|
cilium
GO:0005929
|
1.396 | 7.7e-03 | 146057, 23230, 56123, 2776, 5158, 29922, 283232, 54806, 79738, 56171, 9371, 4131, 79960, 95681, 9738, 56912, 64064, 25790, 8556, 92104, 80705, 2735, 4882, 22873, 2712, 4739, 25876, 79841, 6337, 8452, 80184, 317662, 6787, 11057, 84747, 9231, 27241, 54558, 7840, 4887, 55779, 23639, 92482, 79781, 1235, 79645, 5311, 22863, 57415, 79762, 64792, 151011, 8100, 201163, 481, 57037, 51626, 64927, 9867, 5108, 9940, 9742, 2868, 51314, 22832, 582, 57728, 55064, 79819, 80127, 5577, 585, 80323, 80216, 23116, 4653, 857, 1462, 90355, 79659, 138162, 55112, 55812, 2737, 3800, 65250, 2121, 83450, 54587, 9576, 1811, 114327, 54585, 9863, 8382, 2947, 4117, 6926, 55259, 25924, 80129, 79846, 7802 | |
| Cluster_3 | ||||
|
specific granule
GO:0042581
|
1.628 | 2.9e-03 | 820, 10855, 5004, 4069, 8836, 10321, 3934, 932, 6590, 3608, 1535, 26, 3903, 10562, 6515, 1116, 1991, 1794, 963, 3689, 23250, 8514, 5806, 1088, 2919, 8566, 671, 11322, 945, 5912, 1509, 5329, 221, 1118, 7226, 1536, 5795, 353189, 7133, 1143 | |
|
cytoplasmic vesicle lumen
GO:0060205
|
1.415 | 8.4e-03 | 6280, 6279, 6278, 820, 6286, 10855, 5004, 4069, 8836, 10321, 1521, 3934, 1075, 10549, 11240, 5645, 5709, 51411, 3101, 6590, 6317, 3608, 4860, 2821, 26, 80142, 5713, 2950, 10562, 79980, 51316, 2171, 353, 1116, 6283, 1991, 10576, 1794, 5686, 5695, 5223, 5806, 203068, 5552, 100, 10213, 2919, 2219, 8566, 1476, 2665, 671, 7422, 5315, 5834, 10383 | |
|
secretory granule lumen
GO:0034774
|
1.410 | 1.1e-02 | 6280, 6279, 6278, 820, 6286, 10855, 5004, 4069, 8836, 10321, 1521, 3934, 1075, 10549, 11240, 5645, 5709, 51411, 3101, 6590, 6317, 3608, 4860, 2821, 26, 80142, 5713, 2950, 10562, 79980, 51316, 2171, 353, 1116, 6283, 1991, 10576, 1794, 5686, 5695, 5223, 5806, 203068, 5552, 10213, 2919, 2219, 8566, 1476, 2665, 671, 7422, 5315, 5834, 10383 | |
|
vesicle lumen
GO:0031983
|
1.411 | 1.9e-02 | 6280, 6279, 6278, 820, 6286, 10855, 5004, 4069, 8836, 10321, 1521, 3934, 1075, 10549, 11240, 5645, 5709, 51411, 3101, 6590, 6317, 3608, 4860, 2821, 26, 80142, 5713, 2950, 10562, 79980, 51316, 2171, 353, 1116, 6283, 1991, 10576, 1794, 5686, 5695, 5223, 5806, 203068, 5552, 100, 10213, 2919, 2219, 8566, 1476, 2665, 671, 7422, 5315, 5834, 10383 | |
|
ficolin-1-rich granule lumen
GO:1904813
|
1.517 | 2.9e-02 | 51806, 4318, 10549, 5709, 51411, 3101, 3608, 4860, 2821, 1520, 5713, 2950, 5686, 5695, 5223, 10213, 2219, 56888, 1476, 5315, 4282, 5702, 1509, 10875, 5708, 7415, 5683, 3310, 411, 1460, 5594, 10450, 5718, 3417, 9535, 1512, 11333, 1432, 3326, 5211, 26578, 10097, 51071, 1508, 5236, 29099, 2992, 272, 1522, 10092 | |
|
primary lysosome
GO:0005766
|
1.452 | 2.9e-02 | 6278, 4069, 8836, 1075, 11240, 5645, 6317, 80142, 79980, 51316, 79888, 2171, 9145, 8876, 1991, 10576, 203068, 1088, 2665, 671, 5834, 10383, 9545, 8904, 7415, 6036, 353189, 2588, 3920, 411, 5594, 9588, 5476, 2444, 1650, 719, 10970, 5707, 6809, 10097, 4125, 54472, 23593, 2896, 2760, 84061, 2512, 25801, 4332 | |
|
azurophil granule
GO:0042582
|
1.452 | 2.9e-02 | 6278, 4069, 8836, 1075, 11240, 5645, 6317, 80142, 79980, 51316, 79888, 2171, 9145, 8876, 1991, 10576, 203068, 1088, 2665, 671, 5834, 10383, 9545, 8904, 7415, 6036, 353189, 2588, 3920, 411, 5594, 9588, 5476, 2444, 1650, 719, 10970, 5707, 6809, 10097, 4125, 54472, 23593, 2896, 2760, 84061, 2512, 25801, 4332 | |
After setReadable() was run:
ego3<-ego3 |> setReadable(OrgDb = org.Hs.eg.db,keyType = "ENTREZID")
(EnrichGT(ego3, ClusterNum = 100, P.adj = 1)@gt_object)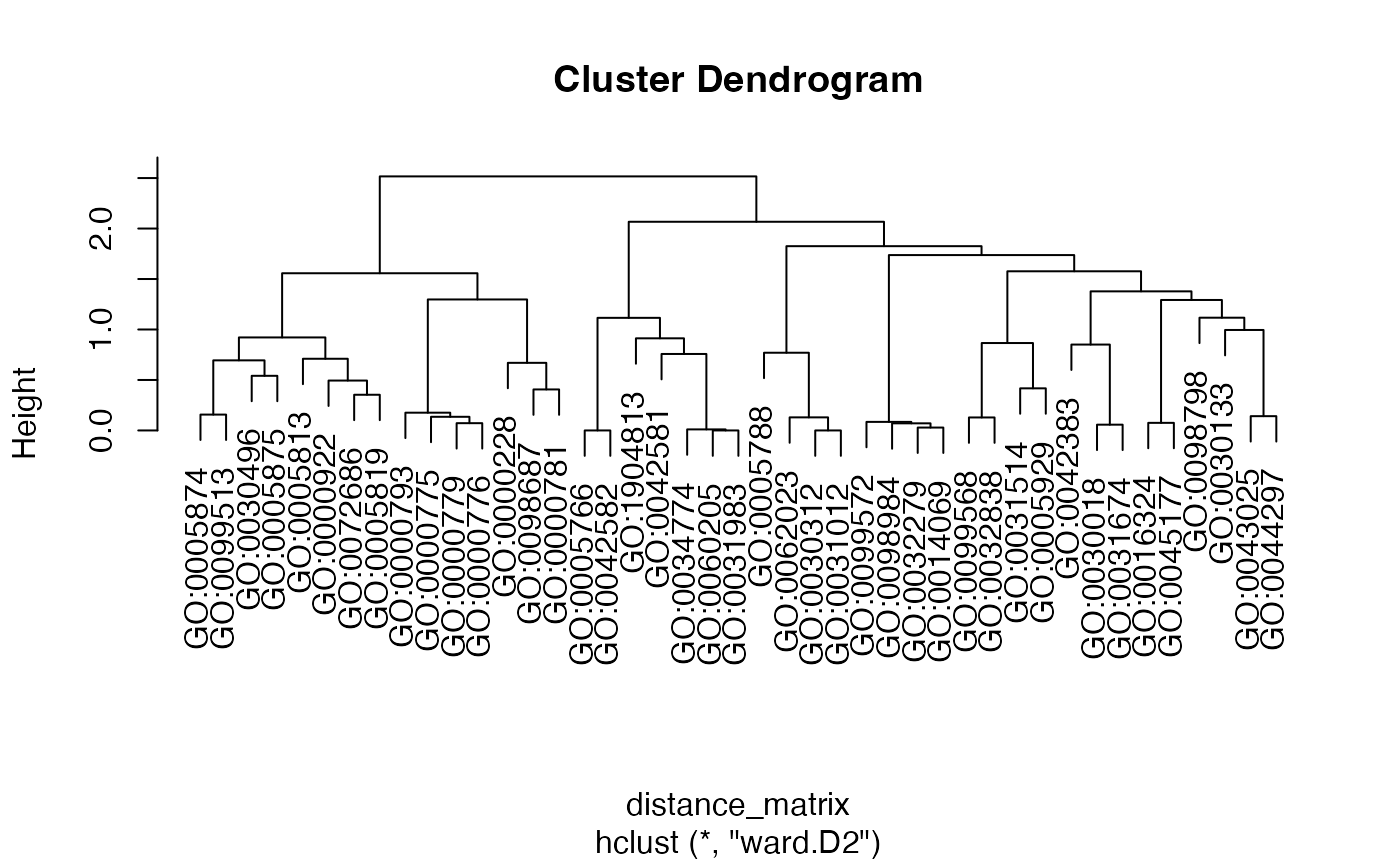
| Parse form: ego3 | ||||
| Split into 3 Clusters. Generated by github@zhimingye/EnrichGT | ||||
| Description | absNES | Padj | core_enrichment | |
|---|---|---|---|---|
| Cluster_1 | ||||
|
chromosome, centromeric region
GO:0000775
|
2.718 | 1.4e-09 | CDCA8, CDC20, CENPE, NDC80, TOP2A, HJURP, SKA1, NEK2, CENPM, CENPN, ERCC6L, MAD2L1, KIF18A, CDT1, BIRC5, EZH2, TTK, NCAPG, AURKB, AURKA, CCNB1, KIF2C, PLK1, BUB1B, ZWINT, CENPU, SPC25, SPAG5, DSCC1, CENPI, OIP5, HELLS, NCAPD2, CENPA, BUB1, CENPF, ZWILCH, CEBPB, H2BC11, DSN1, KNTC1, FIRRM, NCAPD3 | |
|
condensed chromosome, centromeric region
GO:0000779
|
2.713 | 1.4e-09 | CDC20, CENPE, NDC80, HJURP, SKA1, NEK2, CENPM, CENPN, ERCC6L, MAD2L1, KIF18A, CDT1, BIRC5, TTK, NCAPG, AURKB, AURKA, CCNB1, KIF2C, PLK1, BUB1B, ZWINT, CENPU, SPC25, SPAG5, CENPI, NCAPD2, CENPA, BUB1, CENPF, ZWILCH, CEBPB, DSN1, KNTC1, FIRRM, NCAPD3 | |
|
kinetochore
GO:0000776
|
2.670 | 1.4e-09 | CDC20, CENPE, NDC80, HJURP, SKA1, NEK2, CENPM, CENPN, ERCC6L, MAD2L1, KIF18A, CDT1, BIRC5, TTK, AURKB, AURKA, CCNB1, KIF2C, PLK1, BUB1B, ZWINT, CENPU, SPC25, SPAG5, CENPI, BUB1, CENPF, ZWILCH, DSN1, KNTC1, FIRRM | |
|
chromosomal region
GO:0098687
|
2.584 | 1.4e-09 | CDCA8, CDC20, CENPE, NDC80, TOP2A, HJURP, SKA1, NEK2, CENPM, RAD51AP1, CENPN, CDK1, ERCC6L, MAD2L1, KIF18A, CDT1, BIRC5, EZH2, TTK, NCAPG, AURKB, CHEK1, AURKA, CCNB1, MCM5, MCM2, KIF2C, PLK1, BUB1B, ZWINT, CENPU, SPC25, SPAG5, DSCC1, CENPI, RAD51, ORC1, OIP5, HELLS, MCM6, MCM4, FEN1, NCAPD2, CENPA, BUB1, CENPF, PCNA, RECQL4, ZWILCH, MACROH2A2, BLM, DNA2, CEBPB, H2BC11, MCM7, DSN1, MSH2, KNTC1, FIRRM, NCAPD3, HDAC2, PPP1CA, SMC4, MCM3, POLD1, RAD21, CHEK2, ERCC4, ZNF276, SPDL1, SEC13, NBN, MIS18A, ACTL6A, H2BC3, DNMT3A, PARP1, SUV39H1, KDM1A, DNMT1, H2AX, RANGAP1, INCENP, CBX3, CENPQ, POLE3, GAR1, XRCC3 | |
|
nuclear chromosome
GO:0000228
|
2.584 | 1.4e-09 | CDC45, MCM10, TOP2A, NCAPH, NEK2, GINS1, BIRC5, NCAPG, GINS2, CHEK1, MCM5, MCM2, PLK1, RAD51, ORC6, ORC1, MCM6, MCM4, TIPIN, NCAPD2, GINS4, ANP32E, PCNA, GINS3, MACROH2A2, BLM, NCAPG2, POLE2, POLA2, MCM7, SINHCAF, PRIM1, NCAPD3, HDAC2, WDHD1, RUVBL1, SMC4, RCC1, PRIM2, MCM3, POLD1, RAD21, TUBG1, SMC2, TIMELESS, UCHL5, ACTL6A, EXOSC9, PARP1, SUV39H1, RRS1, H2AX, INCENP, POLD2, POLE3, BAZ1A, TTN, RPA3 | |
|
condensed chromosome
GO:0000793
|
2.538 | 1.4e-09 | CDC20, CENPE, NDC80, TOP2A, NCAPH, HJURP, SKA1, NEK2, CENPM, CENPN, ERCC6L, MAD2L1, KIF18A, CDT1, BIRC5, TTK, NCAPG, AURKB, CHEK1, AURKA, CCNB1, KIF2C, PLK1, BUB1B, ZWINT, CENPU, SPC25, SPAG5, CENPI, RAD51, MKI67, NCAPD2, CENPA, BUB1, CENPF, ZWILCH, BLM, CEBPB, NCAPG2, HMGB2, DSN1, KNTC1, FIRRM, NCAPD3, SMC4, RCC1 | |
|
mitotic spindle
GO:0072686
|
2.348 | 1.4e-09 | KIF23, CENPE, ASPM, DLGAP5, SKA1, NUSAP1, TPX2, TACC3, CDK1, MAD2L1, KIF18A, KIF11, TRAT1, AURKB, PRC1, KIFC1, KIF18B, AURKA, CDC6, PLK1, RACGAP1, SPAG5, ECT2, ESPL1, HAUS6, ODAM, GPSM2, PYCR3 | |
|
spindle
GO:0005819
|
2.224 | 1.4e-09 | CDCA8, CDC20, KIF23, CENPE, ASPM, DLGAP5, SKA1, NUSAP1, TPX2, TACC3, NEK2, CDK1, MAD2L1, KIF18A, BIRC5, KIF11, TRAT1, TTK, AURKB, PRC1, KIFC1, KIF18B, KIF20A, AURKA, CCNB1, KIF4A, KIF14, HMMR, KIF2C, SHCBP1, CDC6, PLK1, RACGAP1, BUB1B, SPAG5, ECT2, ESPL1, KIF15, HAUS6, ODAM, GPSM2, CDC25B, PYCR3, CENPF, FBXO5, ZWILCH, SAC3D1, CDC7, DSN1, KIF20B, KNTC1, FIRRM, SLC25A5, PTPN7, CD180, WDR62, BORA, RAD21, TUBB, ACOT13, MICAL3, AUNIP, PSRC1, RMDN1, TUBG1, TUBB3, CIAO2B | |
|
midbody
GO:0030496
|
2.105 | 6.1e-08 | CDCA8, KIF23, CENPE, ASPM, CEP55, NEK2, CDK1, BIRC5, AURKB, PRC1, KIF20A, AURKA, KIF4A, KIF14, SHCBP1, PLK1, RACGAP1, SPAG5, ECT2 | |
|
microtubule
GO:0005874
|
1.708 | 8.3e-06 | KIF23, CENPE, ASPM, SKA1, NUSAP1, TPX2, NEK2, CDK1, KIF18A, BIRC5, KIF11, AURKB, PRC1, KIFC1, KIF18B, KIF20A, AURKA, KIF4A, KIF14, KIF2C, PLK1, RACGAP1, GTSE1, SPAG5, KIF15, HAUS6, MX1, RIBC2, CCT5, TTLL4, NDRG1, ZWILCH, TTLL7, CLIP4, STMN1, TUBA4A, TEKT2, KIF20B, KNTC1, CCT6A, CFAP20, TUBA4B, CCT2, MX2, TUBA1C, KCNAB2, CDK2AP2, TUBB, DNM1L, PSRC1, TCP11L1, RMDN1, TUBG1, TUBB3 | |
| Cluster_2 | ||||
|
collagen-containing extracellular matrix
GO:0062023
|
2.164 | 1.4e-09 | LOXL2, SOD3, COL4A6, NID1, LAMA4, EGFL7, TNC, GDF10, SERPINA3, COL6A1, COL11A1, WNT5A, IGFBP7, THSD4, VWF, COL18A1, CRELD1, CCN2, COL2A1, COL15A1, ADAMTS20, LTBP4, CD151, EDIL3, LTBP3, MGP, WNT2B, PXDN, TGFB2, LAMB1, RARRES2, GPC1, CTSF, SPP2, COL1A2, PRG4, CTSG, SERPINF1, LUM, ANG, EFEMP2, SEMA3B, COL1A1, TIMP3, PRELP, NID2, FBLN5, COL21A1, ANGPTL2, COL6A3, COL8A1, MMRN1, SRPX, ITIH5, CDH13, SHH, MFAP5, PCOLCE, FBLN1, COL3A1, ANGPTL7, COL5A3, COL5A2, WNT5B, MMRN2, THBS2, ADAMTS5, EFEMP1, MMP2, FBLN2, COL7A1, CCN3, MYOC, CXCL12, EMILIN1, HPX, HSPG2, VCAN, COL5A1, COL6A2, LAMA2, LOXL1, MXRA5, LAMA3, LTBP2, SPARC, MEGF9, COL8A2, TGFB1I1, COLQ, BGN, CLU, F13A1, SPON1, GPC4, ECM2, AEBP1, HTRA1, POSTN, DPT, COLEC12, FMOD, MST1, F3, TNN, TGFB3, SERPINA5, CPA3, LAMB2, SPARCL1, COL10A1, FBN1, DCN, LRRC15, BCAM, TPSAB1, COL4A5, AZGP1, THBS4, ELN, COL14A1, COL16A1, ADIPOQ, ANXA9, MMP28, COMP, COL17A1, CILP, NDP, MATN3, TPSB2, ASPN, MFAP4, OGN | |
|
external encapsulating structure
GO:0030312
|
2.105 | 1.4e-09 | CD248, LAMA4, EGFL7, TNC, GDF10, SERPINA3, COL6A1, COL11A1, MMP26, WNT5A, OVGP1, IGFBP7, THSD4, VWF, COL18A1, CRELD1, NDNF, CCN4, CCN2, COL2A1, COL15A1, MMP3, ADAMTS20, LTBP4, CD151, EDIL3, LTBP3, MGP, IGFALS, WNT2B, FGF1, PXDN, OLFML2A, LRRTM4, TGFB2, LAMB1, MMP13, RARRES2, GPC1, CTSF, SPP2, COL1A2, PRG4, CTSG, SERPINF1, LUM, ANG, EFEMP2, SEMA3B, COL1A1, TIMP3, PRELP, NID2, FBLN5, COL21A1, ANGPTL2, COL6A3, COL8A1, MUC6, MMRN1, SRPX, ITIH5, CDH13, SHH, MFAP5, PCOLCE, FBLN1, COL3A1, ANGPTL7, COL5A3, MMP10, COL5A2, WNT5B, OLFML2B, MMRN2, THBS2, ADAMTS5, EFEMP1, MMP2, FBLN2, COL7A1, CCN3, LRIG1, WNT11, MYOC, CXCL12, EMILIN1, HPX, HSPG2, TIMP4, VCAN, COL5A1, COL6A2, LAMA2, LOXL1, MXRA5, LAMA3, LTBP2, CRISPLD2, SPARC, MEGF9, COL8A2, TGFB1I1, COLQ, BGN, CLU, WNT4, F13A1, SPON1, GPC4, ECM2, LRRC32, AEBP1, HTRA1, POSTN, DPT, COLEC12, FMOD, MST1, ANOS1, LRRN3, F3, TNN, TGFB3, SNED1, SERPINA5, CPA3, LAMB2, TNFRSF11B, SPARCL1, COL10A1, FBN1, DCN, LRRC15, BCAM, TPSAB1, COL4A5, AZGP1, THBS4, KERA, ELN, COL14A1, COL16A1, ADIPOQ, FCGBP, ANXA9, MMP28, OMD, COMP, COL17A1, CILP, CCN5, NDP, MATN3, CHAD, TPSB2, ASPN, LRRC17, MFAP4, OGN | |
|
extracellular matrix
GO:0031012
|
2.105 | 1.4e-09 | CD248, LAMA4, EGFL7, TNC, GDF10, SERPINA3, COL6A1, COL11A1, MMP26, WNT5A, OVGP1, IGFBP7, THSD4, VWF, COL18A1, CRELD1, NDNF, CCN4, CCN2, COL2A1, COL15A1, MMP3, ADAMTS20, LTBP4, CD151, EDIL3, LTBP3, MGP, IGFALS, WNT2B, FGF1, PXDN, OLFML2A, LRRTM4, TGFB2, LAMB1, MMP13, RARRES2, GPC1, CTSF, SPP2, COL1A2, PRG4, CTSG, SERPINF1, LUM, ANG, EFEMP2, SEMA3B, COL1A1, TIMP3, PRELP, NID2, FBLN5, COL21A1, ANGPTL2, COL6A3, COL8A1, MUC6, MMRN1, SRPX, ITIH5, CDH13, SHH, MFAP5, PCOLCE, FBLN1, COL3A1, ANGPTL7, COL5A3, MMP10, COL5A2, WNT5B, OLFML2B, MMRN2, THBS2, ADAMTS5, EFEMP1, MMP2, FBLN2, COL7A1, CCN3, LRIG1, WNT11, MYOC, CXCL12, EMILIN1, HPX, HSPG2, TIMP4, VCAN, COL5A1, COL6A2, LAMA2, LOXL1, MXRA5, LAMA3, LTBP2, CRISPLD2, SPARC, MEGF9, COL8A2, TGFB1I1, COLQ, BGN, CLU, WNT4, F13A1, SPON1, GPC4, ECM2, LRRC32, AEBP1, HTRA1, POSTN, DPT, COLEC12, FMOD, MST1, ANOS1, LRRN3, F3, TNN, TGFB3, SNED1, SERPINA5, CPA3, LAMB2, TNFRSF11B, SPARCL1, COL10A1, FBN1, DCN, LRRC15, BCAM, TPSAB1, COL4A5, AZGP1, THBS4, KERA, ELN, COL14A1, COL16A1, ADIPOQ, FCGBP, ANXA9, MMP28, OMD, COMP, COL17A1, CILP, CCN5, NDP, MATN3, CHAD, TPSB2, ASPN, LRRC17, MFAP4, OGN | |
|
endoplasmic reticulum lumen
GO:0005788
|
1.684 | 1.1e-04 | SIL1, CASQ2, TNC, BACE1, COL6A1, COL11A1, MIA3, ESD, WNT5A, FLT3, WFS1, PROC, IGFBP7, COL18A1, ARSJ, COL2A1, COL15A1, FSTL3, CASQ1, SLC27A2, BCHE, LAMB1, HRC, SPP2, COL1A2, ERO1B, EDEM3, GOLM1, ARSD, COL1A1, COL21A1, COL6A3, COL8A1, SHH, COL3A1, COL5A3, FSTL1, COL5A2, WNT5B, ADAMTS5, PDGFC, CHRDL1, COL7A1, GAS6, CST3, VCAN, COL5A1, COL6A2, COL8A2, AFP, CLU, WNT4, ALB, SPON1, PRSS23, MXRA8, LAMB2, IGFBP4, SPARCL1, COL10A1, FBN1, COL4A5, COL14A1, PDGFD, COL16A1, COL17A1, BMP4, MATN3, STC2 | |
|
mitochondrial protein-containing complex
GO:0098798
|
1.574 | 1.6e-03 | PDK1, MRPS17, MRPL15, MRPL13, CYC1, COX5A, MRPL12, MRPS33, SLC25A5, MRPL48, MRPS12, TOMM22, HSD17B10, NDUFS6, NDUFA9, PPIF, DLAT, MRPS7, MRPL42, UQCR10, TIMM17A, UQCRFS1, IDH3A, TIMM44, MRPS15, PDHA1, FXN, CHCHD3, MRPL3, TOMM40, APOO, MCCC1, NDUFS5, ATP5MC3, NDUFV1, ATP5MF, ATP5F1B, COX7B, NDUFB2, GADD45GIP1, TIMM23, NDUFA6, ATP5F1C, IDH3G, MRPS18A, MRPL11, NDUFB11, MRPL2, MPC2, COX8A, PDHA2, MRPL20, COX5B, TIMM8A, PDHX, MRPL9, AGK, TOMM70, TIMM17B, NDUFS1, SAMM50, MFN1, MRPL17, NDUFA4, NDUFA8, MRPL19, DAP3, TIMM8B, ATP5MC1, NDUFA1, COX6B1, CLPX, MRPL33, TIMM13, MRPL18, MRPL4, POLG, SDHA, MRPL35, NDUFB6, SUPV3L1, MRPL39, NDUFS3, MCUB, MRPS11, MRPS2, NDUFB5, PHB2, ATP5PF, COX6A1, UQCRB, NDUFB4, MRPS34, UQCRC1, MRPL44, ATP5PD, ATP5F1E, BAX, VDAC1, MRPL23, PAM16, COX4I1, MRPL58, MRPS16, COX7A2L | |
|
cytoplasmic region
GO:0099568
|
1.638 | 2.0e-03 | DCTN1, SPTBN5, ZC3H14, UNC13A, WDR11, MAP1A, CALY, DNAH2, HSPB1, AP3M2, NME7, MAP2K4, CTNNB1, DNAH7, KIF3B, CFAP45, UNC13B, IFT70A, PCLO, GLI1, SPEF1, PURA, NEK4, PRKCZ, FABP2, TRAK2, PKD2, ATG14, CLASP2, DYNC2LI1, NEFL, IFT140, STXBP1, RAB27B, NME8, NUMA1, CEP162, BBS1, DNAI4, TRAK1, TOGARAM1, DYNC2H1, PIERCE1, SPATA7, GLI3, KIF5C, SYBU, DRC3, SPAG6, EFHC1, MAK, DNAI7, MAPT, DNALI1 | |
|
sarcolemma
GO:0042383
|
1.691 | 2.6e-03 | CACNB2, PPP3CB, KRT19, SGCB, PPP3CA, SCN2A, FLOT1, CAPN3, SLC9A1, COL6A1, UTRN, SLC30A1, PRKG1, SSPN, FLNC, SLC8B1, CASQ1, SGCE, AHNAK, SRI, KCND3, ATP1B1, COL6A3, AQP1, ATP1A2, CACNG4, AHNAK2, SGCD, RTN2, MYOT, IGF1R, ABCC8, CAV1, COL6A2, CACNA1C, LAMA2, SYNC, BGN, TGFB3, FXYD1, CACNA1D | |
|
plasma membrane bounded cell projection cytoplasm
GO:0032838
|
1.607 | 3.4e-03 | WDR11, MAP1A, CALY, DNAH2, HSPB1, AP3M2, NME7, MAP2K4, DNAH7, KIF3B, CFAP45, IFT70A, GLI1, SPEF1, PURA, NEK4, TRAK2, ATG14, DYNC2LI1, NEFL, IFT140, RAB27B, NME8, CEP162, BBS1, DNAI4, TRAK1, TOGARAM1, DYNC2H1, PIERCE1, SPATA7, GLI3, KIF5C, SYBU, DRC3, SPAG6, EFHC1, MAK, DNAI7, MAPT, DNALI1 | |
|
motile cilium
GO:0031514
|
1.566 | 7.1e-03 | DNAH7, IFT46, OXCT2, CFAP45, TSGA10, GK2, SPEF1, SCNN1A, CUL3, ABHD2, SPATA6, CFAP44, DNAAF11, IQCA1, CCR6, PKD2, SEPTIN10, IFT88, ATP1B1, DYNC2LI1, NME8, BBS1, WDR19, SPATA6L, DNAI4, BBS4, DYNC2H1, DRC3, SPAG6, SLC26A3, NME5, GSTM3, MAK, CFAP69, DNALI1 | |
|
cilium
GO:0005929
|
1.396 | 7.7e-03 | TTBK2, VPS13A, PCDHB13, GNAQ, PDE6B, NME7, TMEM80, AHI1, BBS10, DNAH7, KIF3B, MAP1B, JADE1, CEP41, CCP110, IFT46, OXCT2, CFAP45, CDC14A, IFT70A, TSGA10, GLI1, NPR2, DZIP1, GK2, NEDD9, SPEF1, AGBL2, SCNN1A, CUL3, CEP290, FAM149B1, NEK4, ABHD2, UNC119B, DLG5, BBS9, SPATA6, ALMS1, NPY2R, CFAP44, DNAAF11, BBIP1, IQCA1, CCR6, CLXN, PKD2, ATG14, CEP15, C1orf115, IFT22, SEPTIN10, IFT88, FLCN, ATP1B1, ANKMY2, DYNC2LI1, TTC23, PJA2, PCM1, DLEC1, IFT140, GRK4, NME8, CEP162, BBS1, WDR19, SPATA6L, DNAI4, BBOF1, PRKAR2B, BBS4, CCDC68, ALPK1, TOGARAM1, MYOC, CAV1, VCAN, MACIR, DYNC2H1, PIERCE1, DYNC2I1, SPATA7, GLI3, KIF5C, CPLANE1, EVC, DRC3, MXRA8, SPAG6, SLC26A3, EFHC1, LZTFL1, MAGI2, NME5, GSTM3, MAK, TBX3, DNAI7, MYRIP, CCDC170, CFAP69, DNALI1 | |
| Cluster_3 | ||||
|
specific granule
GO:0042581
|
1.628 | 2.9e-03 | CAMP, HPSE, ORM1, LYZ, GGH, CRISP3, LCN2, MS4A3, SLPI, ILF2, CYBA, AOC1, LAIR1, OLFM4, SLC2A3, CHI3L1, ELANE, DOCK2, CD53, ITGB2, ATP11A, KCNAB2, PTX3, CEACAM8, CXCL1, PDXK, BPI, TMC6, CD33, RAP2B, CTSD, PLAUR, ALDH3B1, CHIT1, TRPM2, CYBB, PTPRJ, SLCO4C1, TNFRSF1B, CHRNB4 | |
|
cytoplasmic vesicle lumen
GO:0060205
|
1.415 | 8.4e-03 | S100A9, S100A8, S100A7, CAMP, S100P, HPSE, ORM1, LYZ, GGH, CRISP3, CTSW, LCN2, CTSC, PRDX4, PADI2, PRSS2, PSMD3, BIN2, HK3, SLPI, SERPINB3, ILF2, PNP, GPI, AOC1, PTGES2, PSMD7, GSTP1, OLFM4, DSN1, PLAC8, FABP5, APRT, CHI3L1, S100A12, ELANE, CCT2, DOCK2, PSMA5, PSMB7, PGAM1, PTX3, TUBB, SRGN, ADA, PSMD14, CXCL1, FCN1, PDXK, CSTB, GDI2, BPI, VEGFA, PKM, PYGB, TUBB4B | |
|
secretory granule lumen
GO:0034774
|
1.410 | 1.1e-02 | S100A9, S100A8, S100A7, CAMP, S100P, HPSE, ORM1, LYZ, GGH, CRISP3, CTSW, LCN2, CTSC, PRDX4, PADI2, PRSS2, PSMD3, BIN2, HK3, SLPI, SERPINB3, ILF2, PNP, GPI, AOC1, PTGES2, PSMD7, GSTP1, OLFM4, DSN1, PLAC8, FABP5, APRT, CHI3L1, S100A12, ELANE, CCT2, DOCK2, PSMA5, PSMB7, PGAM1, PTX3, TUBB, SRGN, PSMD14, CXCL1, FCN1, PDXK, CSTB, GDI2, BPI, VEGFA, PKM, PYGB, TUBB4B | |
|
vesicle lumen
GO:0031983
|
1.411 | 1.9e-02 | S100A9, S100A8, S100A7, CAMP, S100P, HPSE, ORM1, LYZ, GGH, CRISP3, CTSW, LCN2, CTSC, PRDX4, PADI2, PRSS2, PSMD3, BIN2, HK3, SLPI, SERPINB3, ILF2, PNP, GPI, AOC1, PTGES2, PSMD7, GSTP1, OLFM4, DSN1, PLAC8, FABP5, APRT, CHI3L1, S100A12, ELANE, CCT2, DOCK2, PSMA5, PSMB7, PGAM1, PTX3, TUBB, SRGN, ADA, PSMD14, CXCL1, FCN1, PDXK, CSTB, GDI2, BPI, VEGFA, PKM, PYGB, TUBB4B | |
|
ficolin-1-rich granule lumen
GO:1904813
|
1.517 | 2.9e-02 | CALML5, MMP9, PRDX4, PSMD3, BIN2, HK3, ILF2, PNP, GPI, CTSS, PSMD7, GSTP1, PSMA5, PSMB7, PGAM1, PSMD14, FCN1, KCMF1, CSTB, PKM, MIF, PSMC3, CTSD, FGL2, PSMD2, VCP, PSMA2, HSPA6, ARSB, CSNK2B, MAPK1, PPIE, PSMD12, IDH1, GMFG, CTSH, PDAP1, MAPK14, HSP90AB1, PFKL, OSTF1, ACTR2, DERA, CTSB, PGM1, COMMD9, GYG1, AMPD3, CTSZ, ARPC5 | |
|
primary lysosome
GO:0005766
|
1.452 | 2.9e-02 | S100A7, LYZ, GGH, CTSC, PADI2, PRSS2, SERPINB3, PTGES2, DSN1, PLAC8, LPCAT1, FABP5, SYNGR1, VNN1, ELANE, CCT2, TUBB, CEACAM8, GDI2, BPI, PYGB, TUBB4B, RAB3D, CPNE1, VCP, RNASE2, SLCO4C1, GALNS, LAMP2, ARSB, MAPK1, PRDX6, CTSA, FRK, DDOST, C3AR1, CKAP4, PSMD1, STX3, ACTR2, MAN2B1, TOLLIP, HEBP2, GRN, GM2A, MAGT1, FTL, GCA, MNDA | |
|
azurophil granule
GO:0042582
|
1.452 | 2.9e-02 | S100A7, LYZ, GGH, CTSC, PADI2, PRSS2, SERPINB3, PTGES2, DSN1, PLAC8, LPCAT1, FABP5, SYNGR1, VNN1, ELANE, CCT2, TUBB, CEACAM8, GDI2, BPI, PYGB, TUBB4B, RAB3D, CPNE1, VCP, RNASE2, SLCO4C1, GALNS, LAMP2, ARSB, MAPK1, PRDX6, CTSA, FRK, DDOST, C3AR1, CKAP4, PSMD1, STX3, ACTR2, MAN2B1, TOLLIP, HEBP2, GRN, GM2A, MAGT1, FTL, GCA, MNDA | |
Session info
sessioninfo::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.3.3 (2024-02-29)
#> os macOS 15.0
#> system aarch64, darwin20
#> ui X11
#> language en
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz Asia/Shanghai
#> date 2024-10-12
#> pandoc 3.1.11 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/aarch64/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> AnnotationDbi * 1.64.1 2023-11-02 [1] Bioconductor
#> ape 5.7-1 2023-03-13 [1] CRAN (R 4.3.0)
#> aplot 0.2.2 2023-10-06 [1] CRAN (R 4.3.1)
#> Biobase * 2.62.0 2023-10-26 [1] Bioconductor
#> BiocGenerics * 0.48.1 2023-11-02 [1] Bioconductor
#> BiocParallel 1.36.0 2023-10-26 [1] Bioconductor
#> Biostrings 2.70.2 2024-01-30 [1] Bioconductor 3.18 (R 4.3.2)
#> bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
#> bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
#> bitops 1.0-7 2021-04-24 [1] CRAN (R 4.3.0)
#> blob 1.2.4 2023-03-17 [1] CRAN (R 4.3.0)
#> bslib 0.6.1 2023-11-28 [1] CRAN (R 4.3.1)
#> cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
#> cli * 3.6.3 2024-06-21 [1] CRAN (R 4.3.3)
#> clusterProfiler * 4.10.1 2024-03-09 [1] Bioconductor 3.18 (R 4.3.3)
#> codetools 0.2-19 2023-02-01 [1] CRAN (R 4.3.3)
#> colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
#> cowplot 1.1.3 2024-01-22 [1] CRAN (R 4.3.1)
#> crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.0)
#> data.table 1.15.4 2024-03-30 [1] CRAN (R 4.3.1)
#> DBI 1.2.2 2024-02-16 [1] CRAN (R 4.3.1)
#> desc 1.4.3 2023-12-10 [1] CRAN (R 4.3.1)
#> digest 0.6.35 2024-03-11 [1] CRAN (R 4.3.1)
#> DOSE 3.28.2 2023-12-12 [1] Bioconductor 3.18 (R 4.3.2)
#> dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.1)
#> EnrichGT * 0.2.8.5 2024-10-12 [1] local
#> enrichplot 1.22.0 2023-11-06 [1] Bioconductor
#> evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.1)
#> fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.1)
#> farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
#> fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
#> fastmatch 1.1-4 2023-08-18 [1] CRAN (R 4.3.0)
#> fgsea 1.28.0 2023-10-26 [1] Bioconductor
#> float 0.3-2 2023-12-10 [1] CRAN (R 4.3.1)
#> fontawesome 0.5.2 2023-08-19 [1] CRAN (R 4.3.0)
#> forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
#> fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.0)
#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
#> GenomeInfoDb 1.38.7 2024-03-09 [1] Bioconductor 3.18 (R 4.3.3)
#> GenomeInfoDbData 1.2.11 2024-03-18 [1] Bioconductor
#> ggforce 0.4.2 2024-02-19 [1] CRAN (R 4.3.1)
#> ggfun 0.1.4 2024-01-19 [1] CRAN (R 4.3.1)
#> ggplot2 * 3.5.0 2024-02-23 [1] CRAN (R 4.3.1)
#> ggplotify 0.1.2 2023-08-09 [1] CRAN (R 4.3.0)
#> ggraph 2.2.1 2024-03-07 [1] CRAN (R 4.3.1)
#> ggrepel 0.9.5 2024-01-10 [1] CRAN (R 4.3.1)
#> ggtree 3.10.1 2024-02-27 [1] Bioconductor 3.18 (R 4.3.2)
#> glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.1)
#> GO.db 3.18.0 2024-03-18 [1] Bioconductor
#> GOSemSim 2.28.1 2024-01-20 [1] Bioconductor 3.18 (R 4.3.2)
#> graphlayouts 1.1.1 2024-03-09 [1] CRAN (R 4.3.1)
#> gridExtra 2.3 2017-09-09 [1] CRAN (R 4.3.0)
#> gridGraphics 0.5-1 2020-12-13 [1] CRAN (R 4.3.0)
#> gson 0.1.0 2023-03-07 [1] CRAN (R 4.3.0)
#> gt * 0.11.0 2024-07-09 [1] CRAN (R 4.3.3)
#> gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
#> HDO.db 0.99.1 2024-03-18 [1] Bioconductor
#> highr 0.10 2022-12-22 [1] CRAN (R 4.3.0)
#> hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
#> htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.1)
#> httr 1.4.7 2023-08-15 [1] CRAN (R 4.3.0)
#> igraph 2.0.3 2024-03-13 [1] CRAN (R 4.3.1)
#> IRanges * 2.36.0 2023-10-26 [1] Bioconductor
#> jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
#> jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.1)
#> KEGGREST 1.42.0 2023-10-26 [1] Bioconductor
#> knitr 1.45 2023-10-30 [1] CRAN (R 4.3.1)
#> lattice 0.22-5 2023-10-24 [1] CRAN (R 4.3.3)
#> lazyeval 0.2.2 2019-03-15 [1] CRAN (R 4.3.0)
#> lgr 0.4.4 2022-09-05 [1] CRAN (R 4.3.0)
#> lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.1)
#> lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.1)
#> magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
#> MASS 7.3-60.0.1 2024-01-13 [1] CRAN (R 4.3.3)
#> Matrix 1.6-5 2024-01-11 [1] CRAN (R 4.3.3)
#> memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
#> mlapi 0.1.1 2022-04-24 [1] CRAN (R 4.3.0)
#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
#> nlme 3.1-164 2023-11-27 [1] CRAN (R 4.3.3)
#> org.Hs.eg.db * 3.18.0 2024-03-18 [1] Bioconductor
#> patchwork 1.2.0 2024-01-08 [1] CRAN (R 4.3.1)
#> pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
#> pkgdown 2.0.7 2022-12-14 [1] CRAN (R 4.3.0)
#> plyr 1.8.9 2023-10-02 [1] CRAN (R 4.3.1)
#> png 0.1-8 2022-11-29 [1] CRAN (R 4.3.0)
#> polyclip 1.10-6 2023-09-27 [1] CRAN (R 4.3.1)
#> proxy 0.4-27 2022-06-09 [1] CRAN (R 4.3.0)
#> purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
#> qvalue 2.34.0 2023-10-26 [1] Bioconductor
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
#> ragg 1.3.0 2024-03-13 [1] CRAN (R 4.3.1)
#> RColorBrewer 1.1-3 2022-04-03 [1] CRAN (R 4.3.0)
#> Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.1)
#> RCurl 1.98-1.14 2024-01-09 [1] CRAN (R 4.3.1)
#> readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.1)
#> reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.3.0)
#> RhpcBLASctl 0.23-42 2023-02-11 [1] CRAN (R 4.3.0)
#> rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.1)
#> rmarkdown 2.26 2024-03-05 [1] CRAN (R 4.3.1)
#> rsparse 0.5.2 2024-06-28 [1] CRAN (R 4.3.3)
#> RSQLite 2.3.5 2024-01-21 [1] CRAN (R 4.3.1)
#> rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.0)
#> S4Vectors * 0.40.2 2023-11-25 [1] Bioconductor 3.18 (R 4.3.2)
#> sass 0.4.9 2024-03-15 [1] CRAN (R 4.3.1)
#> scales 1.3.0 2023-11-28 [1] CRAN (R 4.3.1)
#> scatterpie 0.2.1 2023-06-07 [1] CRAN (R 4.3.0)
#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
#> shadowtext 0.1.3 2024-01-19 [1] CRAN (R 4.3.1)
#> stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.1)
#> stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.1)
#> systemfonts 1.1.0 2024-05-15 [1] CRAN (R 4.3.3)
#> text2vec * 0.6.4 2023-11-09 [1] CRAN (R 4.3.1)
#> textshaping 0.3.7 2023-10-09 [1] CRAN (R 4.3.1)
#> tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
#> tidygraph 1.3.1 2024-01-30 [1] CRAN (R 4.3.1)
#> tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.1)
#> tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.3.1)
#> tidytree 0.4.6 2023-12-12 [1] CRAN (R 4.3.1)
#> tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
#> timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.1)
#> treeio 1.26.0 2023-11-06 [1] Bioconductor
#> tweenr 2.0.3 2024-02-26 [1] CRAN (R 4.3.1)
#> tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
#> utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.1)
#> vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.1)
#> viridis 0.6.5 2024-01-29 [1] CRAN (R 4.3.1)
#> viridisLite 0.4.2 2023-05-02 [1] CRAN (R 4.3.0)
#> withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.1)
#> xfun 0.42 2024-02-08 [1] CRAN (R 4.3.1)
#> xml2 1.3.6 2023-12-04 [1] CRAN (R 4.3.1)
#> XVector 0.42.0 2023-10-26 [1] Bioconductor
#> yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.1)
#> yulab.utils 0.1.4 2024-01-28 [1] CRAN (R 4.3.1)
#> zlibbioc 1.48.0 2023-10-26 [1] Bioconductor
#>
#> [1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
#>
#> ──────────────────────────────────────────────────────────────────────────────