Benchmark of enriched result interpreter (VS Other packages)
c_benchmark.Rmd
library(EnrichGT)
#> Loading required package: dplyr
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
#> Loading required package: tibble
#> Loading required package: gt
#> Loading required package: cli
#>
#> ── EnrichGT ────────────────────────────────────────────────────────────────────
#> ℹ View your enrichment result by entring `EnrichGT(result)`
#> → by Zhiming Ye, https://github.com/ZhimingYe/EnrichGT
suppressMessages({
library(tidyverse)
library(gt)
library(clusterProfiler)
library(org.Hs.eg.db)
library(simplifyEnrichment)
library(aPEAR)
})Benchmark of packages
In the design of EnrichGT, a minimalist approach is
followed, prioritizing the simplest (rather than the most elaborate or
trendy) methods to complete the re-clustering of enrichment analysis
results, even if certain machine learning classifiers might yield better
computational performance. Since this package is primarily targeted at
biologists, the focus is on interpretability rather than computational
superiority.
Compared to well-known packages in the field, such as
simplifyEnrichment and aPEAR, EnrichGT is significantly
faster. While the former two may require over 8 seconds to cluster more
than 1500 GO terms, EnrichGT typically completes the process in around 2
seconds.
simplifyEnrichment
mat = GO_similarity(testWith1700Rows@result$ID)
#> You haven't provided value for `ont`, guess it as `BP`.
start_time <- Sys.time()
df = simplifyGO(mat)
#> Cluster 1709 terms by 'binary_cut'... 33 clusters, used 10.8387 secs.
#> Perform keywords enrichment for 9 GO lists...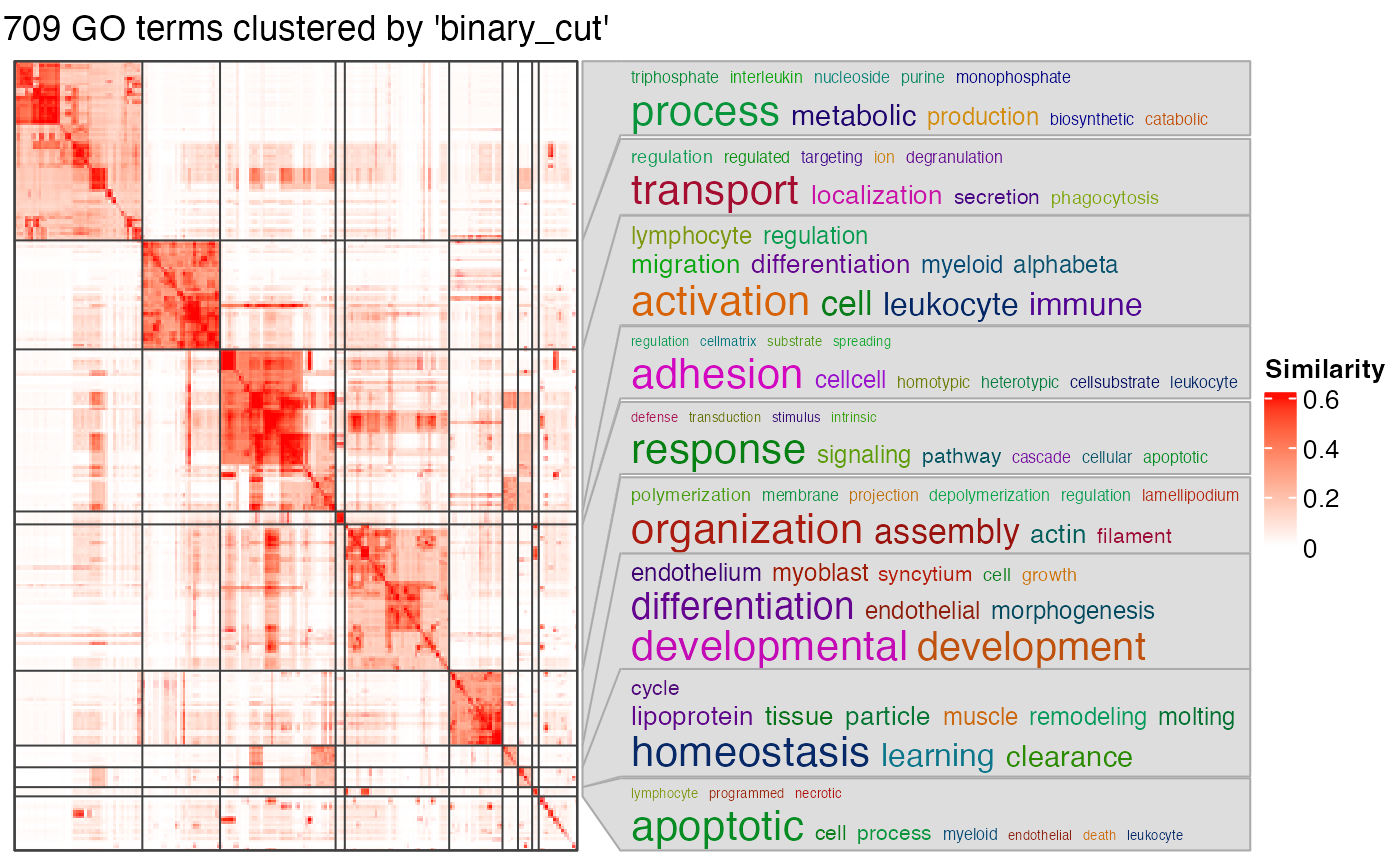
end_time <- Sys.time()
t1<- end_time - start_time
t1
#> Time difference of 1.413719 minsEnrichGT
start_time <- Sys.time()
obj<-testWith1700Rows |> EnrichGT()
#> ℹ =====[SUGGESTION]=====
#> You are passing an object from GO Enrichment.
#> Please ensure that `obj |> clusterProfiler::simplify()` is executed, to pre-simplify result,
#> For better enriched result.
#> Loading required package: text2vec
#>
#> Attaching package: 'text2vec'
#> The following object is masked from 'package:BiocGenerics':
#>
#> normalize
#> ℹ Too many rows! It might be slow...
#> Working, but please consider increase P.adj ...
obj@gt_object| Parse form: testWith1700Rows | ||||
| Split into 17 Clusters. Generated by github@zhimingye/EnrichGT | ||||
| Description | Count | PCT | Padj | geneID |
|---|---|---|---|---|
| Cluster_1 | ||||
|
oxidative phosphorylation
GO:0006119
|
32 | 5.10 | 3.2e-19 | Uqcrq, Ndufa3, Uqcr10, Ndufc1, Ndufs5, Cox6a1, Ndufs8, Atp5pb, Ndufv3, Ndufb9, Cyc1, Iscu, Ndufb1, Ndufs2, Ndufb3, Cox7b, Ndufb10, Mtch2, Ndufa13, Ndufa1, Coa6, Uqcr11, Sdhb, Ndufs3, Ndufa7, Ndufa8, Ndufs6, Cox5b, Cox7c, Uqcc2, Ndufa12, Ndufv2 |
|
aerobic respiration
GO:0009060
|
35 | 5.60 | 1.6e-18 | Uqcrq, Ndufa3, Uqcr10, Ndufc1, Ndufs5, Cox6a1, Ndufs8, Atp5pb, Ndufv3, Ndufb9, Cyc1, Iscu, Ndufb1, Ndufs2, Hif1a, Ndufb3, Cox7b, Suclg1, Ndufb10, Mtch2, Ndufa13, Ndufa1, Fh1, Coa6, Uqcr11, Sdhb, Ndufs3, Ndufa7, Ndufa8, Ndufs6, Cox5b, Cox7c, Uqcc2, Ndufa12, Ndufv2 |
|
cellular respiration
GO:0045333
|
38 | 6.00 | 1.2e-17 | Prelid1, Uqcrq, Ndufa3, Uqcr10, Ndufc1, Etfb, Ndufs5, Cox6a1, Ndufs8, Atp5pb, Ndufv3, Ndufb9, Cyc1, Iscu, Ndufb1, Ndufs2, Hif1a, Ndufb3, Cox7b, Suclg1, Ndufb10, Mtch2, Ndufa13, Ndufa1, Fh1, Coa6, Uqcr11, Cisd1, Sdhb, Ndufs3, Ndufa7, Ndufa8, Ndufs6, Cox5b, Cox7c, Uqcc2, Ndufa12, Ndufv2 |
|
generation of precursor metabolites and energy
GO:0006091
|
49 | 7.80 | 5.5e-16 | App, Prelid1, Uqcrq, Aldoa, Taldo1, Gapdh, Pgd, Ndufa3, Uqcr10, Ldha, Ndufc1, Hdac4, Etfb, Ndufs5, Cox6a1, Ndufs8, Ucp2, Atp5pb, Ndufv3, Ndufb9, Cyc1, Iscu, Ndufb1, Ndufs2, Hif1a, Ndufb3, Cox7b, Suclg1, Ndufb10, Mtch2, Ndufa13, Ndufa1, Ndufa4, Fh1, Coa6, Stk40, Uqcr11, Cisd1, Sdhb, Ndufs3, Ndufa7, Ndufa8, Il6st, Ndufs6, Cox5b, Cox7c, Uqcc2, Ndufa12, Ndufv2 |
|
mitochondrial ATP synthesis coupled electron transport
GO:0042775
|
22 | 3.50 | 1.3e-15 | Uqcrq, Uqcr10, Cox6a1, Ndufs8, Ndufv3, Ndufb9, Cyc1, Iscu, Ndufs2, Cox7b, Mtch2, Coa6, Uqcr11, Sdhb, Ndufs3, Ndufa7, Ndufa8, Ndufs6, Cox5b, Cox7c, Ndufa12, Ndufv2 |
|
energy derivation by oxidation of organic compounds
GO:0015980
|
41 | 6.50 | 1.4e-15 | Prelid1, Uqcrq, Ndufa3, Uqcr10, Ldha, Ndufc1, Etfb, Ndufs5, Cox6a1, Ndufs8, Atp5pb, Ndufv3, Ndufb9, Cyc1, Iscu, Ndufb1, Ndufs2, Hif1a, Ndufb3, Cox7b, Suclg1, Ndufb10, Mtch2, Ndufa13, Ndufa1, Fh1, Coa6, Stk40, Uqcr11, Cisd1, Sdhb, Ndufs3, Ndufa7, Ndufa8, Il6st, Ndufs6, Cox5b, Cox7c, Uqcc2, Ndufa12, Ndufv2 |
|
ATP synthesis coupled electron transport
GO:0042773
|
22 | 3.50 | 1.7e-15 | Uqcrq, Uqcr10, Cox6a1, Ndufs8, Ndufv3, Ndufb9, Cyc1, Iscu, Ndufs2, Cox7b, Mtch2, Coa6, Uqcr11, Sdhb, Ndufs3, Ndufa7, Ndufa8, Ndufs6, Cox5b, Cox7c, Ndufa12, Ndufv2 |
|
proton motive force-driven mitochondrial ATP synthesis
GO:0042776
|
20 | 3.20 | 2.6e-15 | Ndufa3, Ndufc1, Ndufs5, Ndufs8, Atp5pb, Ndufv3, Ndufb9, Ndufb1, Ndufs2, Ndufb3, Ndufb10, Ndufa13, Ndufa1, Sdhb, Ndufs3, Ndufa7, Ndufa8, Ndufs6, Ndufa12, Ndufv2 |
|
ribonucleoside triphosphate biosynthetic process
GO:0009201
|
25 | 4.00 | 3.3e-15 | Nme2, Aldoa, Ndufa3, Ndufc1, Trem2, Uck2, Ndufs5, Ndufs8, Atp5pb, Nme1, Ndufv3, Ndufb9, Ndufb1, Ndufs2, Ndufb3, Ndufb10, Ndufa13, Ndufa1, Sdhb, Ndufs3, Ndufa7, Ndufa8, Ndufs6, Ndufa12, Ndufv2 |
|
nucleoside triphosphate biosynthetic process
GO:0009142
|
25 | 4.00 | 1.0e-14 | Nme2, Aldoa, Ndufa3, Ndufc1, Trem2, Uck2, Ndufs5, Ndufs8, Atp5pb, Nme1, Ndufv3, Ndufb9, Ndufb1, Ndufs2, Ndufb3, Ndufb10, Ndufa13, Ndufa1, Sdhb, Ndufs3, Ndufa7, Ndufa8, Ndufs6, Ndufa12, Ndufv2 |
| Cluster_2 | ||||
|
phagocytosis
GO:0002275
|
35 | 5.60 | 7.7e-16 | Ccr2, Prtn3, Thbs1, Myh9, Bin2, Fcgr2b, Cyba, Cd300lf, Fcgr1, Cfp, Ncf4, Gm5150, Nod2, Msr1, Syk, Itgam, Lrp1, Pycard, Cd302, Siglece, Trem2, Rab7b, Rapgef1, Fgr, Atg3, Rab27a, Anxa11, Ano6, Myd88, Plscr1, Pros1, Lbp, Coro1a, Rac1, Abca1 |
|
regulation of endocytosis
GO:0033003
|
35 | 5.60 | 8.5e-12 | Prtn3, App, Myh9, Fcgr2b, Cyba, Cd300lf, Fcgr1, Cfp, Flot1, Gm5150, Nod2, Picalm, Bmp2k, Syk, Lrp1, Pycard, Ppt1, Apoc2, Siglece, Trem2, Rapgef1, Anxa2, Fgr, Atg3, Rab27a, Iqsec1, Ptpn1, Ano6, Vps28, Plscr1, Pros1, Lbp, Snx9, Rab4b, Lgals3 |
|
regulation of phagocytosis
GO:0042060
|
22 | 3.50 | 9.2e-12 | Prtn3, Myh9, Fcgr2b, Cyba, Cd300lf, Fcgr1, Cfp, Gm5150, Nod2, Syk, Lrp1, Pycard, Siglece, Trem2, Rapgef1, Fgr, Atg3, Rab27a, Ano6, Plscr1, Pros1, Lbp |
|
mitochondrial translation
GO:0043299
|
20 | 3.20 | 7.5e-10 | Mrpl52, Aurkaip1, Mrpl48, Mrpl58, Mrpl14, Mrpl57, Mrpl21, Mrps24, Mrps18c, Mrps15, Mrps21, Mrps34, Mrpl35, Mrps9, Mrps16, Mrpl4, Ndufa7, Uqcc2, Mrpl10, Mrpl34 |
|
mitochondrial gene expression
GO:0045576
|
21 | 3.30 | 9.1e-09 | Mrpl52, Aurkaip1, Mrpl48, Cfh, Mrpl58, Mrpl14, Mrpl57, Mrpl21, Mrps24, Mrps18c, Mrps15, Mrps21, Mrps34, Mrpl35, Mrps9, Mrps16, Mrpl4, Ndufa7, Uqcc2, Mrpl10, Mrpl34 |
|
positive regulation of endocytosis
GO:0050878
|
23 | 3.70 | 1.2e-08 | App, Myh9, Fcgr2b, Cyba, Cd300lf, Fcgr1, Cfp, Flot1, Gm5150, Nod2, Picalm, Syk, Lrp1, Pycard, Ppt1, Trem2, Rapgef1, Anxa2, Rab27a, Ano6, Vps28, Pros1, Lbp |
|
positive regulation of phagocytosis
GO:0043300
|
16 | 2.50 | 1.4e-08 | Myh9, Fcgr2b, Cyba, Cd300lf, Fcgr1, Cfp, Gm5150, Nod2, Lrp1, Pycard, Trem2, Rapgef1, Rab27a, Ano6, Pros1, Lbp |
|
cellular component disassembly
GO:0006887
|
32 | 5.10 | 6.5e-07 | Tspo, Lcp1, Svil, Mid1, Sting1, Flot1, Nod2, Uvrag, Vps13c, Itgam, Ctss, Retreg1, Trem2, Asph, Scaf8, Slc25a4, Mrpl58, Atg3, Gbf1, Sptbn1, Map4k4, Hif1a, Chmp2a, Lamc1, Iqsec1, Cib1, Capg, Capzb, Tomm7, Chmp1a, Cdc37, Htt |
|
response to peptide
GO:0002444
|
32 | 5.10 | 7.1e-07 | Ptpn22, App, Fcgr2b, Gnai2, Ggh, Zfp36l1, Pde3b, Slc39a14, Cyfip1, Nod2, Vps13c, Plcb1, Lrp1, Braf, Ptpre, Eif4ebp1, Trem2, Sort1, Hadha, Cyc1, Adora2b, Socs3, Ednrb, Stxbp3, Sorl1, Slc2a1, Ptpn1, Eif2b2, Pld1, Stat4, Lgmn, Rac1 |
|
leukocyte aggregation
GO:0017157
|
7 | 1.10 | 3.8e-06 | S100a9, S100a8, Thbs1, Cd44, Rac2, Cfh, Msn |
| Cluster_3 | ||||
|
leukocyte migration
GO:0072593
|
42 | 6.70 | 1.2e-14 | Ccr2, S100a9, S100a8, Prtn3, Ptpn22, Thbs1, Sell, C5ar1, App, Ccr1, Bst1, Retnlg, Nod2, Plcb1, Pla2g7, Aimp1, Syk, Itgam, Selplg, Pycard, Rac2, Trem2, P2ry12, Ripor2, Ext1, Mtus1, Trem3, Gbf1, Ednrb, Ano6, Myd88, Cd200r1, Tnfsf14, F11r, Bsg, Lbp, Trem1, Msn, Lgals3, Coro1a, Lgmn, Rac1 |
|
myeloid leukocyte migration
GO:2000379
|
33 | 5.20 | 4.3e-14 | Ccr2, S100a9, S100a8, Prtn3, Thbs1, Sell, C5ar1, App, Ccr1, Bst1, Retnlg, Nod2, Plcb1, Pla2g7, Syk, Itgam, Rac2, Trem2, P2ry12, Ripor2, Mtus1, Trem3, Gbf1, Ednrb, Ano6, Myd88, Cd200r1, Bsg, Lbp, Trem1, Lgals3, Lgmn, Rac1 |
|
leukocyte chemotaxis
GO:0006979
|
29 | 4.60 | 1.5e-11 | Ccr2, S100a9, S100a8, Thbs1, Sell, C5ar1, App, Ccr1, Bst1, Retnlg, Nod2, Pla2g7, Syk, Itgam, Rac2, Ripor2, Mtus1, Trem3, Gbf1, Ednrb, Ano6, Tnfsf14, Bsg, Lbp, Trem1, Lgals3, Coro1a, Lgmn, Rac1 |
|
chemotaxis
GO:2000377
|
40 | 6.30 | 2.4e-11 | Ccr2, S100a9, S100a8, Thbs1, Sell, C5ar1, App, Ccr1, Bin2, Bst1, Retnlg, Nod2, Pla2g7, Syk, Itgam, Lrp1, Sema4b, Gab1, Rac2, Trem2, P2ry12, Ripor2, Mtus1, Cysltr1, Saa3, Trem3, Gbf1, Ednrb, Dock4, Ano6, Tnfsf14, Bsg, Lbp, Cmtm7, Fn1, Trem1, Lgals3, Coro1a, Lgmn, Rac1 |
|
taxis
GO:0051402
|
40 | 6.30 | 2.7e-11 | Ccr2, S100a9, S100a8, Thbs1, Sell, C5ar1, App, Ccr1, Bin2, Bst1, Retnlg, Nod2, Pla2g7, Syk, Itgam, Lrp1, Sema4b, Gab1, Rac2, Trem2, P2ry12, Ripor2, Mtus1, Cysltr1, Saa3, Trem3, Gbf1, Ednrb, Dock4, Ano6, Tnfsf14, Bsg, Lbp, Cmtm7, Fn1, Trem1, Lgals3, Coro1a, Lgmn, Rac1 |
|
cell chemotaxis
GO:0010574
|
33 | 5.20 | 6.0e-11 | Ccr2, S100a9, S100a8, Thbs1, Sell, C5ar1, App, Ccr1, Bin2, Bst1, Retnlg, Nod2, Pla2g7, Syk, Itgam, Gab1, Rac2, Ripor2, Mtus1, Saa3, Trem3, Gbf1, Ednrb, Dock4, Ano6, Tnfsf14, Bsg, Lbp, Trem1, Lgals3, Coro1a, Lgmn, Rac1 |
|
regulation of leukocyte migration
GO:0043523
|
28 | 4.40 | 1.7e-10 | Ccr2, Ptpn22, Thbs1, Sell, C5ar1, App, Ccr1, Bst1, Nod2, Plcb1, Pla2g7, Pycard, Rac2, Trem2, P2ry12, Ripor2, Mtus1, Ano6, Myd88, Cd200r1, Tnfsf14, Lbp, Trem1, Msn, Lgals3, Coro1a, Lgmn, Rac1 |
|
cell activation involved in immune response
GO:0010573
|
31 | 4.90 | 1.8e-09 | Ccr2, Il4ra, App, Lcp1, St3gal1, Milr1, Syk, Itgam, Lrp1, Pycard, Lgals1, Bcl3, Rac2, Trem2, Cd244a, Gab2, Fgr, Gbf1, Rab27a, Adora2b, Clec4e, Apbb1ip, Myd88, Havcr2, Cd180, Lbp, Psen2, Stxbp2, Stat4, Lgals3, Coro1a |
|
neutrophil migration
GO:0050890
|
19 | 3.00 | 1.5e-08 | S100a9, S100a8, Prtn3, Sell, C5ar1, Bst1, Nod2, Syk, Itgam, Rac2, Ripor2, Trem3, Gbf1, Myd88, Bsg, Lbp, Trem1, Lgals3, Rac1 |
|
neutrophil chemotaxis
GO:0010575
|
17 | 2.70 | 2.0e-08 | S100a9, S100a8, Sell, C5ar1, Bst1, Nod2, Syk, Itgam, Rac2, Ripor2, Trem3, Gbf1, Bsg, Lbp, Trem1, Lgals3, Rac1 |
| Cluster_4 | ||||
|
leukocyte cell-cell adhesion
GO:0030099
|
43 | 6.80 | 9.1e-14 | Ccr2, S100a9, S100a8, Il4ra, Ptpn22, Itpkb, Thbs1, Sell, Tarm1, Lrg1, Flot2, Gm5150, Itga5, Syk, Itgam, Selplg, Cd44, Runx1, Pycard, Lgals1, Scgb1a1, Rac2, Bad, Ripor2, Cd244a, Zfp608, Ext1, Cfh, Pag1, Runx3, Tnip1, Havcr2, Gpnmb, Tnfsf14, F11r, Marchf7, Il6st, Arg2, Msn, Tnfaip8l2, Ass1, Lgals3, Coro1a |
|
positive regulation of leukocyte activation
GO:0002573
|
33 | 5.20 | 4.4e-08 | Ccr2, Ctsc, Il4ra, Ptpn22, Mmp8, Itpkb, Thbs1, Flot2, Bst1, Gm5150, Nod2, Pla2g4a, Cd38, Syk, Itgam, Inpp5d, Runx1, Pycard, Lgals1, Trem2, Bad, Cd244a, Gab2, Fgr, Runx3, Adora2b, Myd88, Havcr2, Tnfsf14, Lbp, Il6st, Stxbp2, Coro1a |
|
regulation of inflammatory response
GO:0045638
|
32 | 5.10 | 4.8e-08 | Ccr2, S100a9, S100a8, Ctsc, Mmp8, App, Fcgr2b, Fcgr1, Sting1, Bst1, Nod2, Pik3ap1, Alox5ap, Cd44, Pycard, Lgals1, Ctss, Scgb1a1, Metrnl, Siglece, Trem2, Tnip1, Adora2b, Socs3, Ednrb, Myd88, Cd200r1, Fbxl2, Gpx1, Lbp, Arnt, Tnfaip8l2 |
|
regulation of T cell activation
GO:1902106
|
32 | 5.10 | 4.8e-08 | Ccr2, Il4ra, Ptpn22, Itpkb, Tarm1, Flot2, Gm5150, Prelid1, Syk, Cd44, Runx1, Braf, Pycard, Lgals1, Scgb1a1, Rac2, Bad, Ripor2, Cd244a, Zfp608, Pag1, Runx3, Znhit1, Havcr2, Gpnmb, Tnfsf14, Marchf7, Il6st, Arg2, Tnfaip8l2, Lgals3, Coro1a |
|
immune response-activating signaling pathway
GO:1903707
|
33 | 5.20 | 6.5e-08 | Ptpn22, C5ar1, App, Cyba, Cd300lf, Sting1, Flot1, Kcnn4, Nod2, Cd38, Pik3ap1, Syk, Braf, Pycard, Bmx, Trem2, Rab7b, Rapgef1, Fyb, Tnip1, Clec4e, Traf3, Usp50, Alpk1, Myd88, Havcr2, Eif2b2, Fbxl2, Plscr1, Riok3, Lbp, Psen2, Lgals3 |
|
immune response-regulating signaling pathway
GO:0045637
|
33 | 5.20 | 1.0e-07 | Ptpn22, C5ar1, App, Cyba, Cd300lf, Sting1, Flot1, Kcnn4, Nod2, Cd38, Pik3ap1, Syk, Braf, Pycard, Bmx, Trem2, Rab7b, Rapgef1, Fyb, Tnip1, Clec4e, Traf3, Usp50, Alpk1, Myd88, Havcr2, Eif2b2, Fbxl2, Plscr1, Riok3, Lbp, Psen2, Lgals3 |
|
positive regulation of cell activation
GO:0002762
|
33 | 5.20 | 1.3e-07 | Ccr2, Ctsc, Il4ra, Ptpn22, Mmp8, Itpkb, Thbs1, Flot2, Bst1, Gm5150, Nod2, Pla2g4a, Cd38, Syk, Itgam, Inpp5d, Runx1, Pycard, Lgals1, Trem2, Bad, Cd244a, Gab2, Fgr, Runx3, Adora2b, Myd88, Havcr2, Tnfsf14, Lbp, Il6st, Stxbp2, Coro1a |
|
interleukin-6 production
GO:0002761
|
20 | 3.20 | 2.1e-07 | Ptpn22, App, Cyba, Capn2, Nod2, Syk, Inpp5d, Pycard, Tmem106a, Trem2, Rab7b, Adora2b, Il18rap, Myd88, Havcr2, Cd200r1, Cd300ld, Bsg, Lbp, Psen2 |
|
regulation of innate immune response
GO:0010721
|
33 | 5.20 | 3.5e-07 | Ptpn22, App, Ccr1, Cyba, Cd300lf, Sting1, Flot1, Nod2, Pik3ap1, Serpinb1a, Gfer, Syk, Casp6, Pycard, Trem2, Rab7b, Cfh, Fgr, Tnip1, Trem3, Clec4e, Traf3, Usp50, Il18rap, Alpk1, Myd88, Havcr2, Fbxl2, Plscr1, Riok3, Lbp, Cdc37, Gbp7 |
|
activation of innate immune response
GO:0045671
|
24 | 3.80 | 8.7e-07 | Ptpn22, App, Cyba, Cd300lf, Sting1, Flot1, Nod2, Pik3ap1, Syk, Casp6, Pycard, Trem2, Rab7b, Tnip1, Clec4e, Traf3, Usp50, Alpk1, Myd88, Havcr2, Fbxl2, Riok3, Lbp, Gbp7 |
| Cluster_5 | ||||
|
myeloid leukocyte activation
GO:1904062
|
34 | 5.40 | 1.1e-13 | Ccr2, Pirb, Ctsc, Il4ra, Mmp8, Thbs1, C5ar1, App, Cd300lf, Pla2g4a, Milr1, Crtc3, Syk, Itgam, Pycard, Ptpre, Rac2, Adam9, Tmem106a, Trem2, Ifngr1, Cd244a, Gab2, Fgr, Adora2b, Ndrg1, Il18rap, Myd88, Havcr2, Plscr1, Rbpj, Lbp, Fn1, Stxbp2 |
|
membrane invagination
GO:0051403
|
15 | 2.40 | 2.0e-08 | Thbs1, Myh9, Bin2, Fcgr2b, Fcgr1, Msr1, Itgam, Siglece, Trem2, Chmp2a, Ano6, Lbp, Snx9, Rac1, Abca1 |
|
plasma membrane invagination
GO:0032872
|
14 | 2.20 | 4.8e-08 | Thbs1, Myh9, Bin2, Fcgr2b, Fcgr1, Msr1, Itgam, Siglece, Trem2, Ano6, Lbp, Snx9, Rac1, Abca1 |
|
phagocytosis, engulfment
GO:0070302
|
13 | 2.10 | 8.5e-08 | Thbs1, Myh9, Bin2, Fcgr2b, Fcgr1, Msr1, Itgam, Siglece, Trem2, Ano6, Lbp, Rac1, Abca1 |
|
cytokine-mediated signaling pathway
GO:0010959
|
33 | 5.20 | 1.0e-07 | Ccr2, Pirb, Il4ra, Mmp8, Ifitm2, Ccr1, Cd300lf, Sting1, Plcb1, Syk, Pycard, Gab1, Csf2rb, Il13ra1, Il1r2, Trem2, Hdac4, Bad, Pira12, Ifngr1, Ext1, Ppp2cb, Hif1a, Traf3, Il18rap, Cib1, Myd88, Pira2, Ifitm1, Cdc37, Il6st, Lifr, Stat4 |
|
macrophage activation
GO:0031098
|
15 | 2.40 | 1.2e-06 | Ctsc, Il4ra, Mmp8, Thbs1, C5ar1, App, Pla2g4a, Crtc3, Syk, Itgam, Tmem106a, Trem2, Ifngr1, Havcr2, Lbp |
|
positive regulation of macrophage activation
GO:0006816
|
8 | 1.30 | 9.4e-06 | Ctsc, Il4ra, Mmp8, Thbs1, Pla2g4a, Trem2, Havcr2, Lbp |
|
regulation of lipid transport
GO:1902074
|
16 | 2.50 | 1.5e-05 | Atp8a1, Thbs1, Tspo, Prelid1, Pla2g4a, Syk, Runx1, Lrp1, Apoc2, Trem2, Naxe, Anxa2, Commd1, Sar1b, Map2k6, Abca1 |
|
lipid transport
GO:0010038
|
28 | 4.40 | 2.3e-05 | Atp8a1, Thbs1, Tspo, Kcnn4, Prelid1, Pla2g4a, Atp11a, Vps13c, Msr1, Syk, Runx1, Lrp1, Apoc2, Trem2, Naxe, Anxa2, Commd1, Slc2a1, Ano6, Plscr1, Pitpna, Got2, Sar1b, Map2k6, Lbp, Slc25a11, Esyt1, Abca1 |
|
regulation of lipid localization
GO:0032874
|
17 | 2.70 | 3.3e-05 | Atp8a1, Thbs1, Tspo, Prelid1, Pla2g4a, Msr1, Syk, Runx1, Lrp1, Apoc2, Trem2, Naxe, Anxa2, Commd1, Sar1b, Map2k6, Abca1 |
| Cluster_6 | ||||
|
negative regulation of leukocyte activation
GO:1905517
|
24 | 3.80 | 2.6e-09 | Ccr2, Il4ra, Ptpn22, Tarm1, Fcgr2b, Cd300lf, Milr1, Inpp5d, Cd44, Runx1, Scgb1a1, Tnfrsf13b, Ripor2, Zfp608, Pag1, Fgr, Runx3, Havcr2, Gpnmb, Fn1, Marchf7, Arg2, Tnfaip8l2, Lgals3 |
|
negative regulation of cell activation
GO:1905521
|
25 | 4.00 | 2.8e-09 | Ccr2, Il4ra, Ptpn22, Tarm1, Fcgr2b, Cd300lf, Milr1, Inpp5d, Cd44, Runx1, Scgb1a1, Tnfrsf13b, Trem2, Ripor2, Zfp608, Pag1, Fgr, Runx3, Havcr2, Gpnmb, Fn1, Marchf7, Arg2, Tnfaip8l2, Lgals3 |
|
negative regulation of lymphocyte activation
GO:0043491
|
20 | 3.20 | 1.0e-07 | Il4ra, Ptpn22, Tarm1, Fcgr2b, Inpp5d, Cd44, Runx1, Scgb1a1, Tnfrsf13b, Ripor2, Zfp608, Pag1, Fgr, Runx3, Havcr2, Gpnmb, Marchf7, Arg2, Tnfaip8l2, Lgals3 |
|
regulation of apoptotic signaling pathway
GO:0051896
|
31 | 4.90 | 6.4e-07 | S100a9, S100a8, Ctsc, Thbs1, App, Fcgr2b, Gnai2, Prelid1, Cd44, Pycard, Ctss, Trem2, Bad, Trps1, G0s2, Slc25a4, Runx3, Grina, Hif1a, Lmna, Mtch2, Ndufa13, Ptpn1, Plscr1, Gpx1, Ndufs3, Psen2, Marchf7, Htt, Lgals3, Acaa2 |
|
negative regulation of leukocyte cell-cell adhesion
GO:0043277
|
17 | 2.70 | 2.1e-06 | Il4ra, Ptpn22, Tarm1, Cd44, Runx1, Scgb1a1, Ripor2, Zfp608, Pag1, Runx3, Havcr2, Gpnmb, Marchf7, Arg2, Tnfaip8l2, Ass1, Lgals3 |
|
negative regulation of T cell activation
GO:0045765
|
16 | 2.50 | 2.7e-06 | Il4ra, Ptpn22, Tarm1, Cd44, Runx1, Scgb1a1, Ripor2, Zfp608, Pag1, Runx3, Havcr2, Gpnmb, Marchf7, Arg2, Tnfaip8l2, Lgals3 |
|
negative regulation of cell adhesion
GO:1905523
|
23 | 3.70 | 1.9e-05 | Il4ra, Ptpn22, Thbs1, Tarm1, Gcnt2, Pde3b, Cd44, Runx1, Lrp1, Lgals1, Scgb1a1, Ripor2, Zfp608, Rgcc, Pag1, Runx3, Havcr2, Gpnmb, Marchf7, Arg2, Tnfaip8l2, Ass1, Lgals3 |
|
negative regulation of cell-cell adhesion
GO:0048246
|
18 | 2.90 | 2.8e-05 | Il4ra, Ptpn22, Tarm1, Cd44, Runx1, Scgb1a1, Ripor2, Zfp608, Rgcc, Pag1, Runx3, Havcr2, Gpnmb, Marchf7, Arg2, Tnfaip8l2, Ass1, Lgals3 |
|
regulation of intrinsic apoptotic signaling pathway
GO:1901342
|
17 | 2.70 | 4.1e-05 | S100a9, S100a8, App, Fcgr2b, Cd44, Pycard, Trem2, Bad, Grina, Hif1a, Mtch2, Ndufa13, Ptpn1, Plscr1, Gpx1, Ndufs3, Marchf7 |
|
negative regulation of lymphocyte proliferation
GO:0016525
|
11 | 1.70 | 1.9e-04 | Tarm1, Fcgr2b, Inpp5d, Cd44, Scgb1a1, Tnfrsf13b, Ripor2, Havcr2, Gpnmb, Marchf7, Arg2 |
| Cluster_7 | ||||
|
positive regulation of secretion
GO:0016050
|
35 | 5.60 | 4.2e-09 | S100a9, S100a8, Il4ra, Myh9, Cyba, Kcnn4, Pla2g4a, Serp1, Cd38, Plcb1, Rab8b, Aimp1, Syk, Itgam, Runx1, Lrp1, Adam9, Trem2, Bad, Gab2, Anxa2, Rgcc, Fgr, Rab27a, Adora2b, Hif1a, Chmp2a, Map2k6, Rab3gap1, Bsg, Stxbp2, Kmo, Lgals3, Rac1, Anxa7 |
|
positive regulation of secretion by cell
GO:0071985
|
32 | 5.10 | 1.4e-08 | Il4ra, Myh9, Kcnn4, Pla2g4a, Serp1, Cd38, Plcb1, Rab8b, Aimp1, Syk, Itgam, Runx1, Lrp1, Adam9, Trem2, Bad, Gab2, Anxa2, Rgcc, Fgr, Rab27a, Adora2b, Hif1a, Chmp2a, Map2k6, Rab3gap1, Bsg, Stxbp2, Kmo, Lgals3, Rac1, Anxa7 |
|
ameboidal-type cell migration
GO:0032506
|
30 | 4.80 | 8.2e-06 | S100a9, Thbs1, Myh9, Lcn2, Lrg1, Slc8a1, Actr3, Arhgdib, Sema4b, Braf, Atp2b4, Adam9, P2ry12, Gab2, Rgcc, Adora2b, Map4k4, Hif1a, Ednrb, Arid5b, Iqsec1, Cib1, Pfn1, Gpx1, Bsg, Fn1, Gfus, Coro1a, Lgmn, Rac1 |
|
regulation of cell-substrate adhesion
GO:0007033
|
18 | 2.90 | 8.3e-05 | Thbs1, Gcnt2, Myh9, Bst1, Itga5, Lrp1, Braf, Lgals1, Rac2, Peak1, Map4k4, Cib1, Fmn1, Fn1, Gfus, Trem1, Cass4, Rac1 |
|
response to carbohydrate
GO:0000910
|
17 | 2.70 | 2.1e-04 | Thbs1, Myh9, Zfp36l1, Pde3b, Slc39a14, Plcb1, Casp6, Lrp1, Trem2, Bad, Ucp2, Map4k4, Hif1a, Ern1, Eif2b2, Gpx1, Rac1 |
|
epithelial cell migration
GO:0061756
|
21 | 3.30 | 2.8e-04 | S100a9, Thbs1, Myh9, Lcn2, Lrg1, Atp2b4, Adam9, Gab2, Rgcc, Adora2b, Map4k4, Hif1a, Iqsec1, Cib1, Pfn1, Gpx1, Bsg, Gfus, Coro1a, Lgmn, Rac1 |
|
peptide secretion
GO:0009100
|
21 | 3.30 | 2.8e-04 | S100a9, S100a8, Myh9, Pde3b, Serp1, Cd38, Plcb1, Rab8b, Aimp1, Chd7, Lrp1, Bad, Ucp2, Map4k4, Hif1a, Stxbp3, Htt, Uqcc2, Rac1, Abca1, Anxa7 |
|
positive regulation of epithelial cell migration
GO:0006790
|
14 | 2.20 | 2.9e-04 | Thbs1, Lcn2, Adam9, Gab2, Adora2b, Map4k4, Hif1a, Iqsec1, Cib1, Pfn1, Bsg, Gfus, Lgmn, Rac1 |
|
epithelium migration
GO:0072073
|
21 | 3.30 | 2.9e-04 | S100a9, Thbs1, Myh9, Lcn2, Lrg1, Atp2b4, Adam9, Gab2, Rgcc, Adora2b, Map4k4, Hif1a, Iqsec1, Cib1, Pfn1, Gpx1, Bsg, Gfus, Coro1a, Lgmn, Rac1 |
|
response to monosaccharide
GO:1903510
|
16 | 2.50 | 3.1e-04 | Thbs1, Myh9, Pde3b, Slc39a14, Plcb1, Casp6, Lrp1, Trem2, Bad, Ucp2, Map4k4, Hif1a, Ern1, Eif2b2, Gpx1, Rac1 |
| Cluster_8 | ||||
|
regulation of protein-containing complex assembly
GO:0072006
|
32 | 5.10 | 1.8e-07 | Ptpn22, Gda, Lcp1, Svil, Cyfip1, Arhgap18, Actr3, Syk, Pycard, Eif4ebp1, Trem2, P2ry12, Sptbn1, Mpp7, Sorl1, Usp50, Myd88, Capg, Fbxl2, Pfn1, Capzb, Fmn1, Riok3, Sar1b, Rab3gap1, Epn1, Snx9, Msn, Lgals3, Coro1a, Rac1, Abca1 |
|
lamellipodium organization
GO:0001974
|
12 | 1.90 | 2.6e-05 | Snx1, Cyfip1, Actr3, Cd44, Rac2, Hdac4, P2ry12, Spata13, Snx2, Abi3, Capzb, Rac1 |
|
actin filament polymerization
GO:0009123
|
16 | 2.50 | 3.4e-05 | Svil, Cyfip1, Arhgap18, Actr3, Pycard, Pstpip1, Sptbn1, Arpc4, Capg, Pfn1, Capzb, Fmn1, Snx9, Coro1a, Rac1, Gas7 |
|
positive regulation of protein-containing complex assembly
GO:0009262
|
18 | 2.90 | 3.8e-05 | Ptpn22, Gda, Lcp1, Actr3, Syk, Pycard, P2ry12, Mpp7, Usp50, Myd88, Pfn1, Fmn1, Epn1, Snx9, Msn, Lgals3, Rac1, Abca1 |
|
protein polymerization
GO:0006909
|
20 | 3.20 | 1.2e-04 | Gda, Svil, Cyfip1, Arhgap18, Actr3, Pycard, Pstpip1, Ube2c, Sptbn1, Chmp2a, Arpc4, Capg, Pfn1, Capzb, Fmn1, Snx9, Ccdc57, Coro1a, Rac1, Gas7 |
|
actin polymerization or depolymerization
GO:0030100
|
16 | 2.50 | 1.8e-04 | Svil, Cyfip1, Arhgap18, Actr3, Pycard, Pstpip1, Sptbn1, Arpc4, Capg, Pfn1, Capzb, Fmn1, Snx9, Coro1a, Rac1, Gas7 |
|
regulation of lamellipodium organization
GO:0050764
|
8 | 1.30 | 2.9e-04 | Cyfip1, Actr3, Cd44, Rac2, Hdac4, Abi3, Capzb, Rac1 |
|
actin filament organization
GO:0032543
|
26 | 4.10 | 3.4e-04 | Lcp1, Svil, Myh9, Cyfip1, Arhgap18, Actr3, Braf, Pycard, Rhov, Rac2, Rhou, Pstpip1, Rgcc, Gpr65, Sptbn1, Myo1e, Arpc4, Capg, Pfn1, Capzb, Fmn1, F11r, Snx9, Coro1a, Rac1, Gas7 |
|
regulation of actin filament polymerization
GO:0140053
|
13 | 2.10 | 3.8e-04 | Svil, Cyfip1, Arhgap18, Actr3, Pycard, Sptbn1, Capg, Pfn1, Capzb, Fmn1, Snx9, Coro1a, Rac1 |
|
regulation of lamellipodium assembly
GO:0045807
|
7 | 1.10 | 4.7e-04 | Cyfip1, Actr3, Rac2, Hdac4, Abi3, Capzb, Rac1 |
| Cluster_9 | ||||
|
positive regulation of establishment of protein localization
GO:0050766
|
27 | 4.30 | 1.7e-06 | Ptpn22, App, Myh9, Kcnn4, Serp1, Cd38, Plcb1, Actr3, Itgam, Lrp1, Adam9, Trem2, Bad, Asph, Ergic3, Hif1a, Commd1, Sorl1, Vps28, Cib1, Sar1b, Tomm7, Bsg, Camk1, Rac1, Anxa7, Cct7 |
|
positive regulation of protein transport
GO:0022411
|
26 | 4.10 | 2.4e-06 | Ptpn22, App, Myh9, Kcnn4, Serp1, Cd38, Plcb1, Actr3, Itgam, Lrp1, Adam9, Trem2, Bad, Asph, Ergic3, Hif1a, Commd1, Sorl1, Vps28, Cib1, Sar1b, Tomm7, Bsg, Camk1, Rac1, Anxa7 |
|
protein localization to plasma membrane
GO:1901652
|
20 | 3.20 | 4.2e-04 | Flot2, Flot1, Picalm, Rab8b, Actr3, Lrp1, Atp2b4, Trem2, Anxa2, Fyb, Sptbn1, Abi3, Commd1, Sorl1, Cib1, Camk2g, F11r, Bsg, Lgals3, Rac1 |
|
positive regulation of protein localization to cell periphery
GO:0070486
|
9 | 1.40 | 6.1e-04 | Actr3, Lrp1, Atp2b4, Trem2, Epb41l2, Sptbn1, Commd1, Cib1, Lgals3 |
|
regulation of cellular component size
GO:0050900
|
23 | 3.70 | 6.8e-04 | Svil, Kcnn4, Cyfip1, Arhgap18, Picalm, Actr3, Lamtor4, Lrp1, Sema4b, Pycard, Olfm1, Sptbn1, Ano6, Capg, Eif2b2, Pfn1, Capzb, Fmn1, Fn1, Snx9, Coro1a, Rac1, Anxa7 |
|
regulation of protein localization to plasma membrane
GO:0097529
|
11 | 1.70 | 9.8e-04 | Picalm, Actr3, Lrp1, Atp2b4, Trem2, Sptbn1, Abi3, Commd1, Cib1, Camk2g, Lgals3 |
|
positive regulation of protein localization to plasma membrane
GO:0030595
|
8 | 1.30 | 9.8e-04 | Actr3, Lrp1, Atp2b4, Trem2, Sptbn1, Commd1, Cib1, Lgals3 |
|
regulation of protein localization to cell periphery
GO:0006935
|
12 | 1.90 | 2.1e-03 | Picalm, Actr3, Lrp1, Atp2b4, Trem2, Epb41l2, Sptbn1, Abi3, Commd1, Cib1, Camk2g, Lgals3 |
|
regulation of intracellular protein transport
GO:0042330
|
15 | 2.40 | 2.3e-03 | Ptpn22, Lcp1, Actr3, Itgam, Asph, Ergic3, Commd1, Sorl1, Ptpn1, Cib1, Erlec1, Sar1b, Tomm7, Camk1, Os9 |
|
protein localization to cell periphery
GO:0060326
|
21 | 3.30 | 2.5e-03 | Flot2, Flot1, Picalm, Rab8b, Actr3, Lrp1, Atp2b4, Trem2, Anxa2, Fyb, Epb41l2, Sptbn1, Abi3, Commd1, Sorl1, Cib1, Camk2g, F11r, Bsg, Lgals3, Rac1 |
| Cluster_10 | ||||
|
myeloid cell activation involved in immune response
GO:0002685
|
15 | 2.40 | 2.3e-06 | Ccr2, Il4ra, Milr1, Syk, Itgam, Pycard, Rac2, Trem2, Gab2, Fgr, Adora2b, Myd88, Havcr2, Lbp, Stxbp2 |
|
regulation of mast cell activation
GO:0002263
|
11 | 1.70 | 2.7e-06 | Il4ra, Cd300lf, Milr1, Syk, Ptpre, Rac2, Gab2, Fgr, Adora2b, Plscr1, Stxbp2 |
|
wound healing
GO:1990266
|
25 | 4.00 | 2.4e-05 | Ccr2, S100a9, Arhgap24, Thbs1, Myh9, Lrg1, F13a1, Pla2g4a, Syk, Cd44, P2ry12, Ext1, Cfh, Anxa2, Rab27a, Hif1a, Elk3, Stxbp3, Chmp2a, Ano6, Gpx1, Pros1, F11r, Chmp1a, Fn1 |
|
leukocyte degranulation
GO:0030593
|
12 | 1.90 | 4.1e-05 | Ccr2, Il4ra, Milr1, Syk, Itgam, Rac2, Gab2, Fgr, Rab27a, Adora2b, Stxbp2, Coro1a |
|
mast cell activation
GO:0007159
|
12 | 1.90 | 4.1e-05 | Il4ra, Cd300lf, Milr1, Syk, Ptpre, Rac2, Gab2, Fgr, Adora2b, Ndrg1, Plscr1, Stxbp2 |
|
regulation of body fluid levels
GO:0002696
|
23 | 3.70 | 8.1e-05 | S100a9, Thbs1, Cyba, Gnai2, Kcnn4, F13a1, Pla2g4a, Syk, Xdh, P2ry12, Ext1, Cfh, Nme1, Anxa2, Rab27a, Hif1a, Ednrb, Stxbp3, Aprt, Ano6, Pros1, F11r, Anxa7 |
|
regulation of leukocyte degranulation
GO:0050727
|
9 | 1.40 | 1.0e-04 | Ccr2, Il4ra, Syk, Itgam, Rac2, Gab2, Fgr, Adora2b, Stxbp2 |
|
exocytosis
GO:0050863
|
24 | 3.80 | 1.8e-04 | Ccr2, Il4ra, Myh9, Ccr1, Gnai2, Milr1, Rab8b, Syk, Itgam, Braf, Rac2, Tnfaip2, Gab2, Anxa2, Fgr, Rab27a, Adora2b, Stxbp3, Chmp2a, Rab3ip, Rab3gap1, Sdf4, Stxbp2, Coro1a |
|
myeloid leukocyte mediated immunity
GO:0002757
|
14 | 2.20 | 2.3e-04 | Ccr2, Il4ra, Fcgr1, Milr1, Syk, Itgam, Rac2, Gab2, Fgr, Trem3, Adora2b, Myd88, Stxbp2, Trem1 |
|
regulation of exocytosis
GO:0002764
|
17 | 2.70 | 2.5e-04 | Ccr2, Il4ra, Gnai2, Rab8b, Syk, Itgam, Braf, Rac2, Gab2, Anxa2, Fgr, Rab27a, Adora2b, Stxbp3, Chmp2a, Rab3gap1, Stxbp2 |
| Cluster_11 | ||||
|
reactive oxygen species metabolic process
GO:0050867
|
21 | 3.30 | 1.0e-05 | Thbs1, Tspo, App, Cyba, Gnai2, Lcn2, Ncf4, Syk, Xdh, Itgam, Hdac4, Ucp2, Gbf1, Rab27a, Hif1a, Ndufa13, Prdx1, Gpx1, Romo1, Ndufs3, Arg2 |
|
positive regulation of reactive oxygen species metabolic process
GO:0032635
|
12 | 1.90 | 1.2e-05 | Thbs1, Tspo, App, Cyba, Gnai2, Lcn2, Syk, Xdh, Itgam, Hdac4, Rab27a, Romo1 |
|
response to oxidative stress
GO:0045088
|
25 | 4.00 | 5.3e-05 | Gsr, App, Lcn2, Slc8a1, Cd38, Casp6, Gab1, Adam9, Wrn, Ndufs8, Ucp2, Ppp2cb, 1600014C10Rik, Ndufs2, Hif1a, Mgst1, Prdx1, Camk2g, Psmb5, Gpx1, Romo1, Srxn1, Arnt, Ndufa12, Map3k5 |
|
regulation of reactive oxygen species metabolic process
GO:0002218
|
14 | 2.20 | 3.9e-04 | Thbs1, Tspo, App, Cyba, Gnai2, Lcn2, Syk, Xdh, Itgam, Hdac4, Rab27a, Hif1a, Romo1, Arg2 |
|
neuron apoptotic process
GO:0002274
|
21 | 3.30 | 1.0e-03 | C5ar1, App, Fcgr2b, Lcn2, Casp6, Lrp1, Braf, Ppt1, Gapdh, Retreg1, Trem2, Hdac4, Ucp2, Hif1a, Ube2m, Gpx1, Il6st, Htt, Coro1a, Lgmn, Map3k5 |
|
regulation of vascular endothelial growth factor production
GO:0010324
|
6 | 0.95 | 1.4e-03 | Ccr2, C5ar1, Adora2b, Hif1a, Bsg, Arnt |
|
regulation of neuron apoptotic process
GO:0099024
|
18 | 2.90 | 2.2e-03 | C5ar1, App, Fcgr2b, Lcn2, Casp6, Lrp1, Braf, Ppt1, Retreg1, Trem2, Hdac4, Ucp2, Hif1a, Ube2m, Il6st, Htt, Coro1a, Lgmn |
|
vascular endothelial growth factor production
GO:0006911
|
6 | 0.95 | 2.2e-03 | Ccr2, C5ar1, Adora2b, Hif1a, Bsg, Arnt |
|
cognition
GO:0019221
|
20 | 3.20 | 2.5e-03 | Pirb, Atp8a1, C5ar1, App, Lcn2, Itga5, Cyfip1, Picalm, Plcb1, Chd7, Braf, Ppt1, Trem2, Hif1a, Atxn1, Specc1, Psen2, Grcc10, Htt, Lgmn |
|
positive regulation of vascular endothelial growth factor production
GO:0042116
|
5 | 0.79 | 2.9e-03 | C5ar1, Adora2b, Hif1a, Bsg, Arnt |
| Cluster_12 | ||||
|
myeloid cell differentiation
GO:0043032
|
30 | 4.80 | 1.1e-05 | Pirb, Prtn3, Itpkb, App, Myh9, Ccr1, Nme2, Cd300lf, Zfp36l1, Plcb1, Itgam, Inpp5d, Tmem14c, Runx1, Trem2, Pira12, Ucp2, Rab7b, Gab2, Nme1, Hif1a, Cib1, Myd88, Plscr1, Pira2, Rbpj, Psen2, Sfxn1, Clec2g, Clec5a |
|
myeloid leukocyte differentiation
GO:0032368
|
21 | 3.30 | 2.3e-05 | Pirb, Prtn3, App, Ccr1, Nme2, Cd300lf, Zfp36l1, Plcb1, Itgam, Inpp5d, Runx1, Trem2, Pira12, Ucp2, Gab2, Nme1, Myd88, Pira2, Rbpj, Psen2, Clec2g |
|
negative regulation of myeloid cell differentiation
GO:0006869
|
11 | 1.70 | 2.6e-04 | Pirb, Itpkb, Nme2, Zfp36l1, Inpp5d, Runx1, Pira12, Nme1, Cib1, Pira2, Clec2g |
|
negative regulation of leukocyte differentiation
GO:1905952
|
12 | 1.90 | 3.3e-04 | Pirb, Il4ra, Nme2, Inpp5d, Cd44, Runx1, Pira12, Zfp608, Nme1, Runx3, Pira2, Clec2g |
|
negative regulation of hemopoiesis
GO:0002695
|
12 | 1.90 | 4.7e-04 | Pirb, Il4ra, Nme2, Inpp5d, Cd44, Runx1, Pira12, Zfp608, Nme1, Runx3, Pira2, Clec2g |
|
regulation of myeloid cell differentiation
GO:0050866
|
16 | 2.50 | 1.1e-03 | Pirb, Itpkb, Ccr1, Nme2, Zfp36l1, Itgam, Inpp5d, Runx1, Trem2, Pira12, Rab7b, Nme1, Hif1a, Cib1, Pira2, Clec2g |
|
negative regulation of myeloid leukocyte differentiation
GO:0051250
|
8 | 1.30 | 1.2e-03 | Pirb, Nme2, Inpp5d, Runx1, Pira12, Nme1, Pira2, Clec2g |
|
regulation of myeloid leukocyte differentiation
GO:2001233
|
12 | 1.90 | 1.9e-03 | Pirb, Ccr1, Nme2, Zfp36l1, Itgam, Inpp5d, Runx1, Trem2, Pira12, Nme1, Pira2, Clec2g |
|
negative regulation of cell development
GO:1903038
|
18 | 2.90 | 3.1e-03 | Pirb, Il4ra, Tspo, Nme2, Actr3, Inpp5d, Cd44, Runx1, Sema4b, Trem2, Pira12, Zfp608, Nme1, Runx3, Ednrb, Sorl1, Pira2, Clec2g |
|
negative regulation of osteoclast differentiation
GO:0050868
|
5 | 0.79 | 1.7e-02 | Pirb, Inpp5d, Pira12, Pira2, Clec2g |
| Cluster_13 | ||||
|
regulation of monoatomic cation transmembrane transport
GO:0007162
|
25 | 4.00 | 2.2e-05 | Ptpn22, App, Cyba, Lcn2, Slc8a1, Plcb1, Fxyd5, Chd7, Ctss, Atp2b4, Tspan13, Trem2, Asph, Slc25a4, Iscu, Ubr3, Commd1, Ano6, Ndufa4, Glrx, Sestd1, Psen2, Htt, Gnb5, Coro1a |
|
stress-activated MAPK cascade
GO:0022408
|
19 | 3.00 | 7.2e-05 | Ccr2, Ptpn22, App, Mid1, Fcgr2b, Zfp36l1, Igfbp6, Nod2, Plcb1, Xdh, Pycard, Trem2, Rapgef1, Map4k4, Prdx1, Myd88, Map2k6, Lmnb1, Map3k5 |
|
regulation of stress-activated MAPK cascade
GO:2001242
|
17 | 2.70 | 8.5e-05 | Ccr2, Ptpn22, App, Mid1, Fcgr2b, Igfbp6, Nod2, Plcb1, Xdh, Pycard, Trem2, Rapgef1, Map4k4, Prdx1, Myd88, Lmnb1, Map3k5 |
|
regulation of stress-activated protein kinase signaling cascade
GO:0050672
|
17 | 2.70 | 1.1e-04 | Ccr2, Ptpn22, App, Mid1, Fcgr2b, Igfbp6, Nod2, Plcb1, Xdh, Pycard, Trem2, Rapgef1, Map4k4, Prdx1, Myd88, Lmnb1, Map3k5 |
|
regulation of metal ion transport
GO:0051047
|
27 | 4.30 | 1.1e-04 | Ptpn22, Tspo, Ccr1, Cyba, Gnai2, Lcn2, Slc8a1, Plcb1, Fxyd5, Chd7, Atp2b4, Tspan13, Trem2, P2ry12, Asph, Iscu, Ubr3, Commd1, Sik1, Ano6, Glrx, Sestd1, Psen2, Htt, Gnb5, Lgals3, Coro1a |
|
stress-activated protein kinase signaling cascade
GO:1903532
|
19 | 3.00 | 1.4e-04 | Ccr2, Ptpn22, App, Mid1, Fcgr2b, Zfp36l1, Igfbp6, Nod2, Plcb1, Xdh, Pycard, Trem2, Rapgef1, Map4k4, Prdx1, Myd88, Map2k6, Lmnb1, Map3k5 |
|
calcium ion transport
GO:0001667
|
27 | 4.30 | 1.4e-04 | Ptpn22, Tspo, Ccr1, Cyba, Gnai2, Kcnn4, Slc8a1, Plcb1, Smdt1, Chd7, Slc24a1, Atp2b4, Tspan13, P2ry12, Asph, Tmco1, Cysltr1, Ubr3, Ednrb, Ano6, Camk2g, Sestd1, Psen2, Htt, Gnb5, Lgals3, Coro1a |
|
response to salt
GO:0010810
|
22 | 3.50 | 1.8e-04 | Itpkb, Thbs1, Tspo, Sell, App, Gnai2, Slc8a1, Pla2g4a, Plcb1, Alox5ap, Braf, Atp2b4, Adam9, P2ry12, Asph, Ednrb, Anxa11, Hnrnpd, Htt, Lmnb1, Lgmn, Anxa7 |
|
response to metal ion
GO:0009743
|
21 | 3.30 | 2.1e-04 | Itpkb, Thbs1, App, Pam, Ggh, Pla2g4a, Xdh, Alox5ap, Casp6, Braf, Adam9, Ppp2cb, Asph, Anxa11, Plscr1, Hnrnpd, Fn1, Htt, Ass1, Lgmn, Anxa7 |
|
positive regulation of stress-activated MAPK cascade
GO:0010631
|
13 | 2.10 | 2.6e-04 | Ccr2, App, Mid1, Fcgr2b, Igfbp6, Nod2, Plcb1, Xdh, Pycard, Map4k4, Myd88, Lmnb1, Map3k5 |
| Cluster_14 | ||||
|
macrophage migration
GO:0002790
|
10 | 1.60 | 5.3e-05 | Ccr2, Thbs1, C5ar1, Trem2, P2ry12, Mtus1, Ednrb, Cd200r1, Trem1, Lgals3 |
|
regulation of macrophage migration
GO:0010634
|
8 | 1.30 | 2.3e-04 | Ccr2, Thbs1, C5ar1, Trem2, P2ry12, Mtus1, Cd200r1, Trem1 |
|
protein kinase B signaling
GO:0090132
|
18 | 2.90 | 2.6e-03 | Thbs1, Gcnt2, Zfp36l1, Pde3b, Pik3ap1, Xdh, Trem2, P2ry12, Rapgef1, Gab2, Fgr, Cib1, Fbxl2, Gpx1, Mtdh, Fn1, Cass4, Rac1 |
|
regulation of protein kinase B signaling
GO:0034284
|
16 | 2.50 | 2.9e-03 | Thbs1, Gcnt2, Pde3b, Pik3ap1, Xdh, Trem2, P2ry12, Rapgef1, Fgr, Cib1, Fbxl2, Gpx1, Mtdh, Fn1, Cass4, Rac1 |
|
apoptotic cell clearance
GO:0043254
|
6 | 0.95 | 3.3e-03 | Ccr2, Thbs1, Cd300lf, Lrp1, Trem2, Rac1 |
|
regulation of angiogenesis
GO:0097581
|
17 | 2.70 | 4.4e-03 | Ccr2, Thbs1, C5ar1, Lrg1, Itga5, Pde3b, Runx1, Gab1, Atp2b4, Naxe, Stab1, Rgcc, Cysltr1, Hif1a, Epn1, Mtdh, Lgals3 |
|
positive regulation of macrophage migration
GO:0030041
|
5 | 0.79 | 4.8e-03 | Thbs1, C5ar1, Trem2, P2ry12, Trem1 |
|
macrophage chemotaxis
GO:0031334
|
6 | 0.95 | 4.9e-03 | Thbs1, C5ar1, Mtus1, Ednrb, Trem1, Lgals3 |
|
regulation of vasculature development
GO:0051258
|
17 | 2.70 | 4.9e-03 | Ccr2, Thbs1, C5ar1, Lrg1, Itga5, Pde3b, Runx1, Gab1, Atp2b4, Naxe, Stab1, Rgcc, Cysltr1, Hif1a, Epn1, Mtdh, Lgals3 |
|
negative regulation of angiogenesis
GO:0008154
|
8 | 1.30 | 1.7e-02 | Ccr2, Thbs1, Pde3b, Atp2b4, Naxe, Stab1, Rgcc, Epn1 |
| Cluster_15 | ||||
|
vesicle organization
GO:1902743
|
20 | 3.20 | 1.8e-03 | Pla2g4a, Uvrag, Picalm, Rab8b, Sort1, Rab7b, Gbf1, Rab27a, Trappc2l, Chmp2a, Usp50, Atp6v1g1, Trappc6a, Trappc1, Sar1b, Atp6v1e1, Chmp1a, Rnasek, Coro1a, Abca1 |
|
multivesicular body sorting pathway
GO:0007015
|
5 | 0.79 | 7.5e-03 | Sort1, Rab27a, Chmp2a, Vps28, Chmp1a |
|
cytokinetic process
GO:0030833
|
5 | 0.79 | 1.3e-02 | Myh9, Chmp2a, Anxa11, Chmp1a, Snx9 |
|
vacuole organization
GO:0010591
|
12 | 1.90 | 2.2e-02 | Sting1, Ppt1, Rab3gap2, Rab7b, Ext1, Man2a1, Atg3, Chmp2a, Rab3gap1, Chmp1a, Coro1a, Abca1 |
|
cytokinesis
GO:1904951
|
10 | 1.60 | 4.2e-02 | Myh9, Uvrag, Actr3, Pstpip1, Sptbn1, Chmp2a, Anxa11, Chmp1a, Snx9, Dctn3 |
| Cluster_16 | ||||
|
leukocyte adhesion to vascular endothelial cell
GO:0051222
|
6 | 0.95 | 8.8e-03 | Ccr2, Sell, Lrg1, Itgam, Selplg, Ext1 |
|
glycoprotein metabolic process
GO:0072659
|
17 | 2.70 | 1.5e-02 | Gcnt2, Itm2b, St3gal1, Plcb1, Tmem106a, Ext1, Man2a1, Dpm3, Tnip1, Hif1a, Galns, Alg5, Xylt1, Tm9sf2, Chst11, Ost4, Man1a |
|
sulfur compound metabolic process
GO:1904377
|
14 | 2.20 | 3.3e-02 | Xdh, Ext1, Ciao2a, Iscu, Ednrb, Dpep2, Galns, Suclg1, Nfu1, Mpc1, Xylt1, Tm9sf2, Chst11, Acaa2 |
|
kidney epithelium development
GO:0032535
|
9 | 1.40 | 3.7e-02 | Vcan, Pbx1, Cd44, Ext1, Ednrb, Myo1e, Fmn1, Basp1, Arg2 |
|
mucopolysaccharide metabolic process
GO:1903076
|
6 | 0.95 | 4.0e-02 | Cd44, Ext1, Ednrb, Galns, Xylt1, Chst11 |
|
nephron development
GO:1903078
|
9 | 1.40 | 4.5e-02 | Pbx1, Cd44, Sec61a1, Ext1, Cfh, Ednrb, Myo1e, Fmn1, Basp1 |
|
blood vessel remodeling
GO:1904375
|
5 | 0.79 | 5.0e-02 | Ccr2, Chd7, Lrp1, Ext1, Rbpj |
| Cluster_17 | ||||
|
nucleoside monophosphate metabolic process
GO:0033157
|
6 | 0.95 | 2.0e-02 | Gda, Nme2, Xdh, Uck2, Nme1, Aprt |
|
deoxyribonucleotide metabolic process
GO:1990778
|
5 | 0.79 | 2.1e-02 | Gda, Nme2, Xdh, Dera, Nme1 |
end_time <- Sys.time()
t2 <-end_time - start_time
t2
#> Time difference of 1.763355 secsaPEAR
start_time <- Sys.time()
p <- enrichmentNetwork(testWith1700Rows@result, drawEllipses = TRUE, fontSize = 2.5,plotOnly = F)
#> Warning in geom_point(data = coordinates, aes(x = x, y = y, ID = ID, color =
#> color, : Ignoring unknown aesthetics: ID, Cluster, and
#> Cluster size
#> Warning in geom_text(data = labels, aes(x = x, y = y, label = label), size =
#> fontSize, : Ignoring unknown parameters: `segment.size`
p[["plot"]]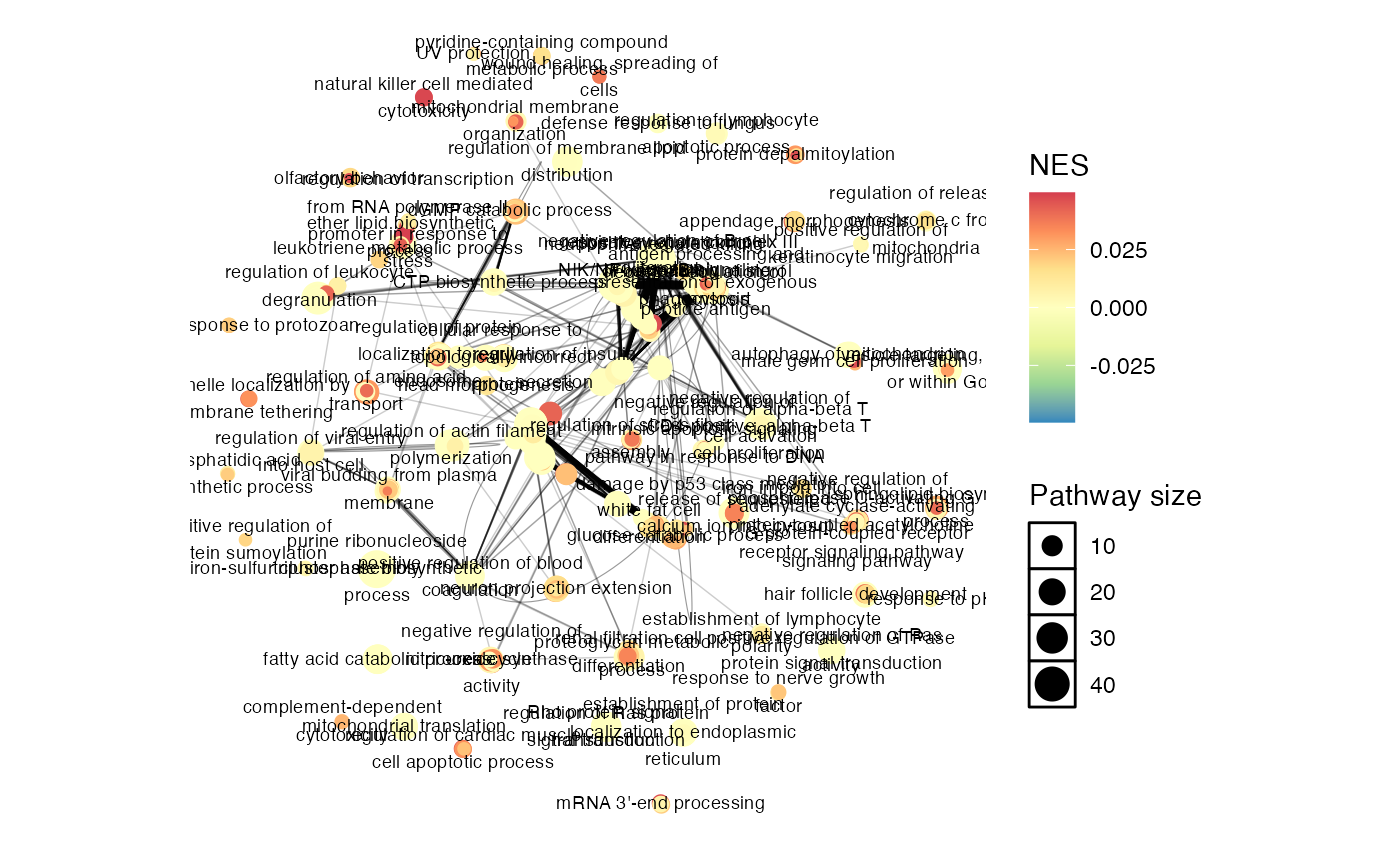
end_time <- Sys.time()
t3 <-end_time - start_time
t3
#> Time difference of 5.709913 minsCompare Calculation Times
You can see the EnrichGT() is the fastest, only using
~2s. simplifyEnrichment takes ~10s, and aPEAR
use almost 5 mins…
Session info
sessioninfo::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.3.3 (2024-02-29)
#> os macOS 15.0
#> system aarch64, darwin20
#> ui X11
#> language en
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz Asia/Shanghai
#> date 2024-10-12
#> pandoc 3.1.11 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/aarch64/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> AnnotationDbi * 1.64.1 2023-11-02 [1] Bioconductor
#> ape 5.7-1 2023-03-13 [1] CRAN (R 4.3.0)
#> aPEAR * 1.0 2024-03-18 [1] Github (ievaKer/aPEAR@4eb4aec)
#> aplot 0.2.2 2023-10-06 [1] CRAN (R 4.3.1)
#> arules 1.7-7 2023-11-29 [1] CRAN (R 4.3.1)
#> bayesbio 1.0.0 2016-05-24 [1] CRAN (R 4.3.0)
#> Biobase * 2.62.0 2023-10-26 [1] Bioconductor
#> BiocGenerics * 0.48.1 2023-11-02 [1] Bioconductor
#> BiocParallel 1.36.0 2023-10-26 [1] Bioconductor
#> Biostrings 2.70.2 2024-01-30 [1] Bioconductor 3.18 (R 4.3.2)
#> bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
#> bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
#> bitops 1.0-7 2021-04-24 [1] CRAN (R 4.3.0)
#> blob 1.2.4 2023-03-17 [1] CRAN (R 4.3.0)
#> bslib 0.6.1 2023-11-28 [1] CRAN (R 4.3.1)
#> cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
#> circlize 0.4.16 2024-02-20 [1] CRAN (R 4.3.1)
#> cli * 3.6.3 2024-06-21 [1] CRAN (R 4.3.3)
#> clue 0.3-65 2023-09-23 [1] CRAN (R 4.3.1)
#> cluster 2.1.6 2023-12-01 [1] CRAN (R 4.3.3)
#> clusterProfiler * 4.10.1 2024-03-09 [1] Bioconductor 3.18 (R 4.3.3)
#> codetools 0.2-19 2023-02-01 [1] CRAN (R 4.3.3)
#> colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
#> ComplexHeatmap 2.18.0 2023-10-26 [1] Bioconductor
#> cowplot 1.1.3 2024-01-22 [1] CRAN (R 4.3.1)
#> crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.0)
#> data.table 1.15.4 2024-03-30 [1] CRAN (R 4.3.1)
#> DBI 1.2.2 2024-02-16 [1] CRAN (R 4.3.1)
#> desc 1.4.3 2023-12-10 [1] CRAN (R 4.3.1)
#> digest 0.6.35 2024-03-11 [1] CRAN (R 4.3.1)
#> doParallel 1.0.17 2022-02-07 [1] CRAN (R 4.3.0)
#> DOSE 3.28.2 2023-12-12 [1] Bioconductor 3.18 (R 4.3.2)
#> dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.1)
#> EnrichGT * 0.2.8.5 2024-10-12 [1] local
#> enrichplot 1.22.0 2023-11-06 [1] Bioconductor
#> evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.1)
#> expm 0.999-9 2024-01-11 [1] CRAN (R 4.3.1)
#> fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.1)
#> farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
#> fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
#> fastmatch 1.1-4 2023-08-18 [1] CRAN (R 4.3.0)
#> fgsea 1.28.0 2023-10-26 [1] Bioconductor
#> float 0.3-2 2023-12-10 [1] CRAN (R 4.3.1)
#> forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
#> foreach 1.5.2 2022-02-02 [1] CRAN (R 4.3.0)
#> fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.0)
#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
#> GenomeInfoDb 1.38.7 2024-03-09 [1] Bioconductor 3.18 (R 4.3.3)
#> GenomeInfoDbData 1.2.11 2024-03-18 [1] Bioconductor
#> GetoptLong 1.0.5 2020-12-15 [1] CRAN (R 4.3.0)
#> ggforce 0.4.2 2024-02-19 [1] CRAN (R 4.3.1)
#> ggfun 0.1.4 2024-01-19 [1] CRAN (R 4.3.1)
#> ggplot2 * 3.5.0 2024-02-23 [1] CRAN (R 4.3.1)
#> ggplotify 0.1.2 2023-08-09 [1] CRAN (R 4.3.0)
#> ggraph 2.2.1 2024-03-07 [1] CRAN (R 4.3.1)
#> ggrepel 0.9.5 2024-01-10 [1] CRAN (R 4.3.1)
#> ggtree 3.10.1 2024-02-27 [1] Bioconductor 3.18 (R 4.3.2)
#> GlobalOptions 0.1.2 2020-06-10 [1] CRAN (R 4.3.0)
#> glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.1)
#> GO.db 3.18.0 2024-03-18 [1] Bioconductor
#> GOSemSim 2.28.1 2024-01-20 [1] Bioconductor 3.18 (R 4.3.2)
#> graphlayouts 1.1.1 2024-03-09 [1] CRAN (R 4.3.1)
#> gridExtra 2.3 2017-09-09 [1] CRAN (R 4.3.0)
#> gridGraphics 0.5-1 2020-12-13 [1] CRAN (R 4.3.0)
#> gson 0.1.0 2023-03-07 [1] CRAN (R 4.3.0)
#> gt * 0.11.0 2024-07-09 [1] CRAN (R 4.3.3)
#> gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
#> HDO.db 0.99.1 2024-03-18 [1] Bioconductor
#> highr 0.10 2022-12-22 [1] CRAN (R 4.3.0)
#> hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
#> htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.1)
#> httr 1.4.7 2023-08-15 [1] CRAN (R 4.3.0)
#> igraph 2.0.3 2024-03-13 [1] CRAN (R 4.3.1)
#> IRanges * 2.36.0 2023-10-26 [1] Bioconductor
#> iterators 1.0.14 2022-02-05 [1] CRAN (R 4.3.0)
#> jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
#> jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.1)
#> KEGGREST 1.42.0 2023-10-26 [1] Bioconductor
#> knitr 1.45 2023-10-30 [1] CRAN (R 4.3.1)
#> labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
#> lattice 0.22-5 2023-10-24 [1] CRAN (R 4.3.3)
#> lazyeval 0.2.2 2019-03-15 [1] CRAN (R 4.3.0)
#> lgr 0.4.4 2022-09-05 [1] CRAN (R 4.3.0)
#> lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.1)
#> lsa 0.73.3 2022-05-09 [1] CRAN (R 4.3.0)
#> lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.1)
#> magick 2.8.4 2024-07-14 [1] CRAN (R 4.3.3)
#> magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
#> MASS 7.3-60.0.1 2024-01-13 [1] CRAN (R 4.3.3)
#> Matrix 1.6-5 2024-01-11 [1] CRAN (R 4.3.3)
#> matrixStats 1.2.0 2023-12-11 [1] CRAN (R 4.3.1)
#> MCL 1.0 2015-03-11 [1] CRAN (R 4.3.0)
#> memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
#> mlapi 0.1.1 2022-04-24 [1] CRAN (R 4.3.0)
#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
#> nlme 3.1-164 2023-11-27 [1] CRAN (R 4.3.3)
#> NLP 0.3-0 2024-08-05 [1] CRAN (R 4.3.3)
#> org.Hs.eg.db * 3.18.0 2024-03-18 [1] Bioconductor
#> patchwork 1.2.0 2024-01-08 [1] CRAN (R 4.3.1)
#> pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
#> pkgdown 2.0.7 2022-12-14 [1] CRAN (R 4.3.0)
#> plyr 1.8.9 2023-10-02 [1] CRAN (R 4.3.1)
#> png 0.1-8 2022-11-29 [1] CRAN (R 4.3.0)
#> polyclip 1.10-6 2023-09-27 [1] CRAN (R 4.3.1)
#> proxy 0.4-27 2022-06-09 [1] CRAN (R 4.3.0)
#> proxyC 0.4.1 2024-04-07 [1] CRAN (R 4.3.1)
#> purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
#> qvalue 2.34.0 2023-10-26 [1] Bioconductor
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
#> ragg 1.3.0 2024-03-13 [1] CRAN (R 4.3.1)
#> RColorBrewer 1.1-3 2022-04-03 [1] CRAN (R 4.3.0)
#> Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.1)
#> RCurl 1.98-1.14 2024-01-09 [1] CRAN (R 4.3.1)
#> readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.1)
#> reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.3.0)
#> RhpcBLASctl 0.23-42 2023-02-11 [1] CRAN (R 4.3.0)
#> rjson 0.2.21 2022-01-09 [1] CRAN (R 4.3.0)
#> rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.1)
#> rmarkdown 2.26 2024-03-05 [1] CRAN (R 4.3.1)
#> rsparse 0.5.2 2024-06-28 [1] CRAN (R 4.3.3)
#> RSQLite 2.3.5 2024-01-21 [1] CRAN (R 4.3.1)
#> rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.0)
#> S4Vectors * 0.40.2 2023-11-25 [1] Bioconductor 3.18 (R 4.3.2)
#> sass 0.4.9 2024-03-15 [1] CRAN (R 4.3.1)
#> scales 1.3.0 2023-11-28 [1] CRAN (R 4.3.1)
#> scatterpie 0.2.1 2023-06-07 [1] CRAN (R 4.3.0)
#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
#> shadowtext 0.1.3 2024-01-19 [1] CRAN (R 4.3.1)
#> shape 1.4.6.1 2024-02-23 [1] CRAN (R 4.3.1)
#> simplifyEnrichment * 1.12.0 2023-10-26 [1] Bioconductor
#> slam 0.1-53 2024-09-02 [1] CRAN (R 4.3.3)
#> SnowballC 0.7.1 2023-04-25 [1] CRAN (R 4.3.0)
#> stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.1)
#> stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.1)
#> systemfonts 1.1.0 2024-05-15 [1] CRAN (R 4.3.3)
#> text2vec * 0.6.4 2023-11-09 [1] CRAN (R 4.3.1)
#> textshaping 0.3.7 2023-10-09 [1] CRAN (R 4.3.1)
#> tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
#> tidygraph 1.3.1 2024-01-30 [1] CRAN (R 4.3.1)
#> tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.1)
#> tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.3.1)
#> tidytree 0.4.6 2023-12-12 [1] CRAN (R 4.3.1)
#> tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
#> timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.1)
#> tm 0.7-14 2024-08-13 [1] CRAN (R 4.3.3)
#> treeio 1.26.0 2023-11-06 [1] Bioconductor
#> tweenr 2.0.3 2024-02-26 [1] CRAN (R 4.3.1)
#> tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
#> utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.1)
#> vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.1)
#> viridis 0.6.5 2024-01-29 [1] CRAN (R 4.3.1)
#> viridisLite 0.4.2 2023-05-02 [1] CRAN (R 4.3.0)
#> withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.1)
#> xfun 0.42 2024-02-08 [1] CRAN (R 4.3.1)
#> xml2 1.3.6 2023-12-04 [1] CRAN (R 4.3.1)
#> XVector 0.42.0 2023-10-26 [1] Bioconductor
#> yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.1)
#> yulab.utils 0.1.4 2024-01-28 [1] CRAN (R 4.3.1)
#> zlibbioc 1.48.0 2023-10-26 [1] Bioconductor
#>
#> [1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
#>
#> ──────────────────────────────────────────────────────────────────────────────